Difference between revisions of "Modules:RegisterImagesBatch-Documentation-3.4"
(→Known bugs: No space in path) |
|||
| Line 72: | Line 72: | ||
===Known bugs=== | ===Known bugs=== | ||
| + | Don't use paths with space characters. | ||
| − | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | + | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. |
===Usability issues=== | ===Usability issues=== | ||
Revision as of 19:41, 12 May 2009
Home < Modules:RegisterImagesBatch-Documentation-3.4Return to Slicer 3.4 Documentation
Module Name
Register Images BatchMake
General Information
Module Type & Category
Type: CLI
Category: Batch Processing
Authors, Collaborators & Contact
- Authors: Julien Finet, Stephen Aylward
- Contact: julien.finet at kitware.com
Module Description
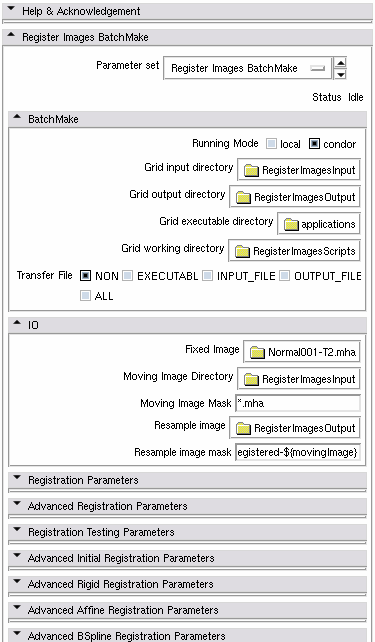
This Module is the BatchMake version of Register Images module, an integrated framework providing access to ITK registration technologies. N moving images can be registered with a single fixed image on a computational grid. The moving images are the images filenames that match a mask in a directory.
Usage
Fixed Image = the fixed image
Moving Image Directory = the directory the moving images share with each others
Moving Image Mask = a mask (concatenated to the Moving Image Directoy) that is used to select only the moving images. If the moving images are in sub directories, separate the sub directories with the '/' character.
Resampled Image Directory = the output directory where the registered images will be saved
Resampled Image Mask = the name of the resampled images created by Register Images. To ensure uniqueness of the name, you can use the ${movingImage} BatchMake variable that contains the name of each moving image.
For the other parameters, please see the Register Images module documentation.
Examples, Use Cases & Tutorials
For the following file tree:
- Images
- Target.mha
- Patient1
- 00001.txt
- 00001.mha
- Patient2
- 00001.txt
- 00001.mha
- Patient3
- 00001.txt
- 00001.mha
- Result
Parameters could be:
Fixed Image: "Images/Target.mha"
Moving Image Directory: "Images"
Moving Image Mask: "Patient*/*.mha"
Resampled Image Directory: "Result"
Resampled Image Mask: "Resampled-{movingImage}"
RegisterImages would then be run 3 times with the following parameters:
RegisterImages ... Images/Target.mha Images/Patient1/00001.mha --resampledImage Resampled-Patient1/00001.mha
RegisterImages ... Images/Target.mha Images/Patient2/00001.mha --resampledImage Resampled-Patient2/00001.mha
RegisterImages ... Images/Target.mha Images/Patient3/00001.mha --resampledImage Resampled-Patient3/00001.mha
Quick Tour of Features and Use
To run the module on a condor grid, open the BatchMake parameter group and choose "condor" as running mode. Set the Input, Output, Executable and Working directories.
Input directory = Moving Image directory (probably on the network)
Output directory = Resampled Image directory (probably on the network)
Executable directory = directory where all your applications are: bmGridStore, bmGridSend, RegisterImages...). This directory shall be the same for all of your condor machines.
Working directory = directory where the BatchMake and condor scripts will be written (probably on the network)
Development
Dependencies
Register Images module
Known bugs
Don't use paths with space characters.
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code: BatchMakeModule.cxx
XML Description: RegisterImagesBatchMakeModule.xml
Documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.