Modules:PETCTFusion-Documentation-3.6
Return to Slicer 3.6 Documentation
PET/CT Fusion Module
General Information
Module Type & Category
Type: Interactive
Category: Base
Authors, Collaborators & Contact
- Author1: Wendy Plesniak, BWH
- Contributor1: Jeffrey Yap, Dana Farber Cancer Institute
- Contributor2: Ron Kikinis, BWH
- Contact: Wendy Plesniak, wjp@bwh.harvard.edu
Module Description
This module computes Standardized Uptake Value (SUV) based on patient body weight, using a PET image and one or more manually drawn VOIs. SUV-bw for VOIs of different color (id) will be assessed separately.
Usage
Use Cases, Examples
This module can be used to quantify tumor response to treatment. Typically, an initial PET/CT study is done prior to treatment, and a followup study is registered to it. ROIs can be specified on the initial study and used to compute SUVmax-bw and SUVmean-bw on both studies.
Tutorials
Links to tutorials explaining how to use this module:
Quick Tour of Features and Use
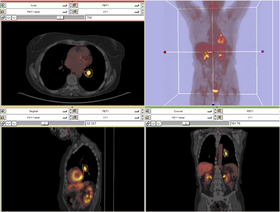
A list panels in the interface, their features, what they mean, and how to use them. For instance:
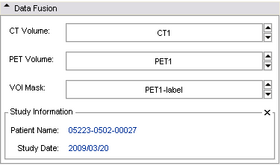
Data Fusion & Study Information:
In the Data Fusion panel shown below, a CT volume and corresponding PET volume should be selected. A pre-created tumor mask (label map) should be selected. This mask can be created in the editor module using the PET volume as a reference volume. This module will compute a separate SUV-bw for each label in the map. So if there are multiple volumes of interest for which individual SUV's should be computed, a separate label (color) should be used for each VOI. Volumes contained within same-label colors will be computed together.
This panel also displays some basic study information, including patient name and study date, as these may be important visual checks for a user.
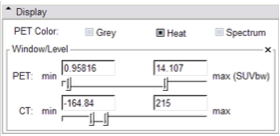
Display panel:
In the module's Display panel shown below, options to select a colorizing scheme for the PET dataset are provided:
- Grey will provide white to black colorization, with black indicating the highest count values.
- Heat will provide a warm color scale, with Dark red lowest, and white the highest count values.
- Spectrum will provide a warm color scale that goes cooler (dark blue) on the low-count end to white at the highest.
This panel also provides a means to adjust the window and level of both PET and CT volumes.
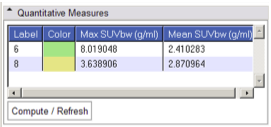
Quantitative Measures panel:
This display panel has the "Compute/Refresh" button which may be selected after datasets are selected. Once SUV has been computed, the panel above will be populated by results for each label in the tumor mask, and indexed by both ID number and color.
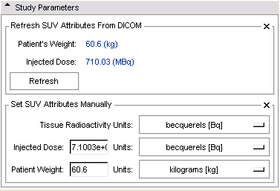
Study Parameters panel:
This panel can be consulted to view other study parameters that may be of interest. Parameters may also be entered manually here, if no DICOM metadata is available for them (note, this feature has not been thoroughly tested.)
Development
Notes from the Developer(s)
This module uses ITK's gdcm to access DICOM metadata used for computing SUV-bw. Philips datasets are not currently handled.
Dependencies
Slicer Core.
Known bugs
Philips PET datasets use private DICOM tags that are not supported by this version of the module. The module should not be used to analyze datasets acquired by Philips systems.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Files:
- vtkPETCTFusion.h
- vtkPETCTFusionLogic.h
- vtkPETCTFusionLogic.cxx
- vtkPETCTFusionGUI.h
- vtkPETCTFusionGUI.cxx
- vtkMRMLPETCTFusionNode.h
- vtkMRMLPETCTFusionNode.cxx
Doxygen documentation:
Acknowledgment
Development of this module was supported by
- The Harvard Clinical and Translational Science Center
- The National Alliance for Medical Image Computing
- The Neuroimage Analysis Center
- The National Center for Image-Guided Therapy, and
- The Surgical Planning Laboratory at Brigham and Women's Hospital.
This module was developed by Wendy Plesniak, Jeffrey Yap and Ron Kikinis