Difference between revisions of "Modules:MRIBiasFieldCorrection-Documentation-3.5"
Sylvainjaume (talk | contribs) (add usage instructions) |
Sylvainjaume (talk | contribs) (move instructions to the usage section) |
||
| Line 6: | Line 6: | ||
===MRI Bias Field Correction=== | ===MRI Bias Field Correction=== | ||
Filtering:MRIBiasFieldCorrection | Filtering:MRIBiasFieldCorrection | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{| | {| | ||
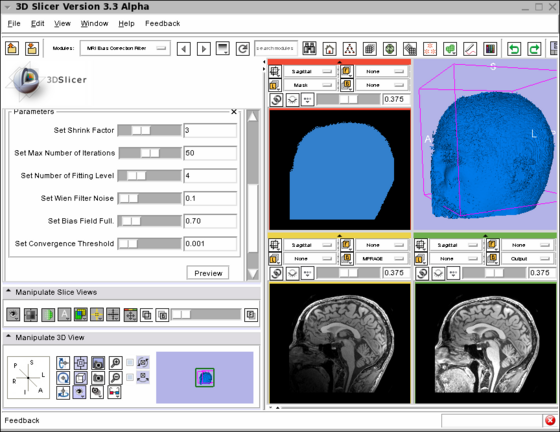
| − | |[[Image:MRI_Bias_Field_Correction_Slicer3.png|thumb|560px|Screenshot of the MRIBiasFieldCorrection module in Slicer3. | + | |[[Image:MRI_Bias_Field_Correction_Slicer3.png|thumb|560px|Screenshot of the MRIBiasFieldCorrection module in Slicer3 during development. |
The screenshot illustrates the application of the MRI Bias Field Correction on the SPL Atlas. | The screenshot illustrates the application of the MRI Bias Field Correction on the SPL Atlas. | ||
| + | Note that the interface was simplified for the production version. | ||
The top left window shows the mask used to defined the ROI where the correction will be applied. | The top left window shows the mask used to defined the ROI where the correction will be applied. | ||
The top right window shows the rendering of the 3D model created using the mask. | The top right window shows the rendering of the 3D model created using the mask. | ||
| Line 65: | Line 46: | ||
== Usage == | == Usage == | ||
| − | * Load the input dataset (Add Volume) | + | * Load the input dataset (Main menu: Add Volume) |
| − | * | + | * Create a Mask Volume using the Editor (Modules > Editor) (the Threshold tool should give a good result) |
| − | * Select the | + | * Select the MRIBiasFieldCorrection module (Modules > Filtering > MRIBiasFieldCorrection) |
| − | * | + | * In the left panel, select the Input Volume |
| − | * Modify the | + | * Select the Mask Volume |
| − | * Click on Apply | + | * In the Preview Volume menu , select 'Create New Volume' |
| − | + | * Do the same for the Output Volume menu | |
| + | * Modify the parameter values if desired (default values gave good results during our experiments) | ||
| + | * Click on Apply | ||
| + | |||
| + | It took 32 min to process a 512x512x30 MRI volume on a MacBook laptop. | ||
===Examples, Use Cases & Tutorials=== | ===Examples, Use Cases & Tutorials=== | ||
Revision as of 00:06, 18 August 2009
Home < Modules:MRIBiasFieldCorrection-Documentation-3.5Return to Slicer 3.5 Documentation
MRI Bias Field Correction
Filtering:MRIBiasFieldCorrection
This module enables the user to get a fast preview of the Bias Field Correction applied on a 2D slice. The advantage of this preview is to try different parameters and visualize the effect in real time.
A Command Line version of this module has been developed to enable the Slicer3 Testing framework. Hence this module is tested during every Nightly build and the result is reported in the Slicer3 Dashboard.
General Information
Module Type & Category
Type: Interactive or CLI
Category: Filtering
Authors, Collaborators & Contact
- Nicolas Rannou: Harvard Medical School, Brigham and Women's Hospital
- Sylvain Jaume: MIT Computer Science and Artificial Intelligence Laboratory
- Contact: <nrannou at bwh.harvard.edu>, <sylvain at csail.mit.edu>
Module Description
The module filters the image to remove the intensity inhomogeneity due to the MRI image acquisition.
Usage
- Load the input dataset (Main menu: Add Volume)
- Create a Mask Volume using the Editor (Modules > Editor) (the Threshold tool should give a good result)
- Select the MRIBiasFieldCorrection module (Modules > Filtering > MRIBiasFieldCorrection)
- In the left panel, select the Input Volume
- Select the Mask Volume
- In the Preview Volume menu , select 'Create New Volume'
- Do the same for the Output Volume menu
- Modify the parameter values if desired (default values gave good results during our experiments)
- Click on Apply
It took 32 min to process a 512x512x30 MRI volume on a MacBook laptop.
Examples, Use Cases & Tutorials
- Note use cases for which this module is especially appropriate, and/or link to examples. (to be defined)
- Link to examples of the module's use (to be defined)
- Link to any existing tutorials (to be defined)
Quick Tour of Features and Use
List all the panels in your interface, their features, what they mean, and how to use them. For instance:
- Input panel:
- Parameters panel:
- Output panel:
- Viewing panel:
Development
Dependencies
No other modules or packages are required to use this module.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
More Information
Acknowledgment
This work has been supported by NA-MIC Algorithms.
References
- A Nonparametric Method for Automatic Correction of Intensity Nonuniformity in MRI Data, J.G. Sled, A.P. Zijdenbos, and A.C. Evans, IEEE Transactions on Medical Imaging, 17(1):87–97, Feb 1998.
- Parametric Estimate of Intensity Inhomogeneities Applied to MRI, M. Styner, C. Brechbhler, G. Szekely, and G. Gerig, IEEE Transactions on Medical Imaging, 19(3):153–165, Mar 2000.
- N4ITK: Nick's N3 ITK Implementation For MRI Bias Field Correction, Tustison N., Gee J., Insight Journal, 2009.