Difference between revisions of "Modules:FuzzySegmentationModule"
(Created page with 'Return to Slicer 3.6 Documentation __NOTOC__ ===Module Name=== Fuzzy Tissue Classification {| border="0" |thumb|280px|Module UI | …') |
|||
| (14 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
| style="text-align: center;" | | | style="text-align: center;" | | ||
{| border="0" | {| border="0" | ||
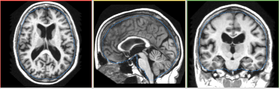
| − | |[[Image:FCMInput-3-6.png|thumb| | + | |[[Image:FCMInput-3-6.png|thumb|280px|Input Image with Mask]] |
|- | |- | ||
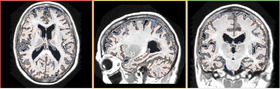
| − | |[[Image:FCMResult-3-6.png|thumb| | + | |[[Image:FCMResultLabels-3-6.png|thumb|280px|Segmentation Results]] |
| + | |- | ||
| + | |[[Image:FCMResult-3-6.png|thumb|280px|Segmentation Results]] | ||
|} | |} | ||
|} | |} | ||
| Line 27: | Line 29: | ||
===Module Description=== | ===Module Description=== | ||
| + | |||
| + | This module computes voxel by voxel tissue classification of an MR brain image using a fuzzy c-means algortihm. Bias field is modeled as a lower order polynomial. Bias field and tissue classification are estimated iteratively in an EM fashion. Internally, each voxel is assigned tissue membership function values, which range from 0 to 1. At any voxel, the sum of membership function of all classes is either 0 (outside of brain), or 1. The membership functions are converted in tissue labels to generate hard segmentation. | ||
== Usage == | == Usage == | ||
===Examples, Use Cases & Tutorials=== | ===Examples, Use Cases & Tutorials=== | ||
| + | |||
| + | This module is typically used to assign tissue labels to an image. For example, in MR brain image analysis, many applications need to know which voxels belong to gray matter, white matter, and CSF. | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| + | * '''Input/Output panels:''' | ||
| + | |||
| + | [[Image:FCM-IOPanels-3-6.png|thumb|right|280px|Input/Output panels]] | ||
| + | This module takes two input volumes and generate two output volumes. '''Input Volume''' is the MR image to be segmented. '''Input Mask''' gives the mask within which segmentation is applied. '''Hard Segmentation''' is an output label map. And '''Bias Field''' is the estimated bias field. | ||
| + | |||
| + | * '''Parameters panel:''' | ||
| + | [[Image:FCM-ParametersPanels-3-6.png|thumb|right|280px|Parameters panels]] | ||
| + | |||
| + | User can specify two parameters: '''NumberOfClasses''' tells the algorithm how many different tissues there are in the given mask. For brain, we use 3 -- gray matter, white matter, and CSF; '''BiasOption''' specifies how bias field is modeled -- 0: no bias correction; 1: global linear; 2: global quadratic; 3: region based linear; 4: region based quadratic. From 0 to 4, computation time increases as the accuracy increases. | ||
== Development == | == Development == | ||
===Dependencies=== | ===Dependencies=== | ||
| + | |||
| + | No other modules are required for this module. The mask can be generated using any skull stripping algorithm. | ||
===Known bugs=== | ===Known bugs=== | ||
| Line 44: | Line 61: | ||
===Usability issues=== | ===Usability issues=== | ||
| − | + | Developed and tested on T1 weighted images. Performance on images from other modality or with other MR contrasts may be limited. | |
===Source code & documentation=== | ===Source code & documentation=== | ||
| − | Source Code: | + | Source Code: [http://viewvc.slicer.org/viewcvs.cgi/trunk/BrainTissueClassification/TissueClassification.cxx?rev=6314&root=NAMICSandBox&view=log TissueClassification.cxx] |
| + | |||
| + | XML Description: [http://viewvc.slicer.org/viewcvs.cgi/trunk/BrainTissueClassification/TissueClassification.xml?rev=6314&root=NAMICSandBox&view=log TissueClassification.xml] | ||
| − | + | Test: [http://viewvc.slicer.org/viewcvs.cgi/trunk/BrainTissueClassification/Testing/TissueClassificationTest.cxx?rev=6314&root=NAMICSandBox&sortby=author&view=log TissueClassificationTest.cxx] | |
Usage: | Usage: | ||
<pre> | <pre> | ||
| + | USAGE: | ||
| + | |||
| + | ../Slicer3-ext/BrainTissueClassification-build/lib/Slicer3/Plugins/Tissu | ||
| + | eClassification | ||
| + | [--returnparameterfile | ||
| + | <std::string>] | ||
| + | [--processinformationaddress | ||
| + | <std::string>] [--xml] [--echo] [-b | ||
| + | <int>] [-c <int>] [--] [--version] | ||
| + | [-h] <std::string> <std::string> | ||
| + | <std::string> <std::string> | ||
| + | |||
| + | |||
| + | Where: | ||
| + | |||
| + | --returnparameterfile <std::string> | ||
| + | Filename in which to write simple return parameters (int, float, | ||
| + | int-vector, etc.) as opposed to bulk return parameters (image, | ||
| + | geometry, transform, measurement, table). | ||
| + | |||
| + | --processinformationaddress <std::string> | ||
| + | Address of a structure to store process information (progress, abort, | ||
| + | etc.). (default: 0) | ||
| + | |||
| + | --xml | ||
| + | Produce xml description of command line arguments (default: 0) | ||
| + | |||
| + | --echo | ||
| + | Echo the command line arguments (default: 0) | ||
| + | |||
| + | -b <int>, --biasoption <int> | ||
| + | Option for bias correction (0: no bias correction; 1: global linear; | ||
| + | 2: global quadratic; 3: region based linear; 4: region based | ||
| + | quadratic) (default: 0) | ||
| + | |||
| + | -c <int>, --class <int> | ||
| + | Number of classes (default: 3) | ||
| + | |||
| + | --, --ignore_rest | ||
| + | Ignores the rest of the labeled arguments following this flag. | ||
| + | |||
| + | --version | ||
| + | Displays version information and exits. | ||
| + | |||
| + | -h, --help | ||
| + | Displays usage information and exits. | ||
| + | |||
| + | <std::string> | ||
| + | (required) Input T1 Image. | ||
| + | |||
| + | <std::string> | ||
| + | (required) Only voxels inside the mask are classified | ||
| + | |||
| + | <std::string> | ||
| + | (required) Output brain mask map. | ||
| + | |||
| + | <std::string> | ||
| + | (required) Estimated bias field | ||
| + | |||
| + | |||
| + | Description: This module computes voxel by voxel tissue classification | ||
| + | of an MR brain image using a fuzzy c-means algortihm. Bias field is | ||
| + | modeled as a lower order polynomial. Bias field and tissue | ||
| + | classification are estimated iteratively in an EM fashion. Internally, | ||
| + | each voxel is assigned tissue membership function values, which range | ||
| + | from 0 to 1. At any voxel, the sum of membership function of all classes | ||
| + | is either 0 (outside of brain), or 1. The membership functions are | ||
| + | converted in tissue labels to generate hard segmentation. | ||
| + | |||
| + | Author(s): Xiaodong Tao, taox @ research . ge . com | ||
| + | |||
| + | Acknowledgements: This work is part of the National Alliance for Medical | ||
| + | Image Computing (NAMIC), funded by the National Institutes of Health | ||
| + | through the NIH Roadmap for Medical Research, Grant U54 EB005149. | ||
| + | Implementation of the Fuzzy Classification was contributed by Dr. | ||
| + | Ming-Ching Chang from GE Research. | ||
| + | |||
</pre> | </pre> | ||
| Line 59: | Line 155: | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
| + | This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Implementation of the Fuzzy Classification was contributed by Dr. Ming-Ching Chang from GE Research. | ||
| + | |||
| + | [http://www.oasis-brains.org/ OASIS datasets] were used to generate images on this page. | ||
===References=== | ===References=== | ||
| + | |||
| + | * Ming-ching Chang, Xiaodong Tao “Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion”, SPIE Medical Imaging, San Diego, CA, 2010. | ||
Latest revision as of 15:08, 30 April 2010
Home < Modules:FuzzySegmentationModuleReturn to Slicer 3.6 Documentation
Module Name
Fuzzy Tissue Classification
|
|
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Author: Xiaodong Tao
- Contact: taox at research.ge.com
Module Description
This module computes voxel by voxel tissue classification of an MR brain image using a fuzzy c-means algortihm. Bias field is modeled as a lower order polynomial. Bias field and tissue classification are estimated iteratively in an EM fashion. Internally, each voxel is assigned tissue membership function values, which range from 0 to 1. At any voxel, the sum of membership function of all classes is either 0 (outside of brain), or 1. The membership functions are converted in tissue labels to generate hard segmentation.
Usage
Examples, Use Cases & Tutorials
This module is typically used to assign tissue labels to an image. For example, in MR brain image analysis, many applications need to know which voxels belong to gray matter, white matter, and CSF.
Quick Tour of Features and Use
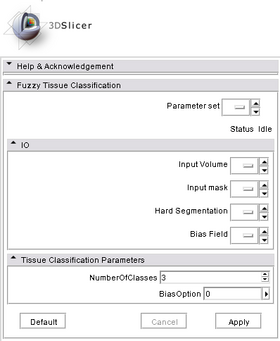
- Input/Output panels:
This module takes two input volumes and generate two output volumes. Input Volume is the MR image to be segmented. Input Mask gives the mask within which segmentation is applied. Hard Segmentation is an output label map. And Bias Field is the estimated bias field.
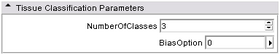
- Parameters panel:
User can specify two parameters: NumberOfClasses tells the algorithm how many different tissues there are in the given mask. For brain, we use 3 -- gray matter, white matter, and CSF; BiasOption specifies how bias field is modeled -- 0: no bias correction; 1: global linear; 2: global quadratic; 3: region based linear; 4: region based quadratic. From 0 to 4, computation time increases as the accuracy increases.
Development
Dependencies
No other modules are required for this module. The mask can be generated using any skull stripping algorithm.
Known bugs
None.
Usability issues
Developed and tested on T1 weighted images. Performance on images from other modality or with other MR contrasts may be limited.
Source code & documentation
Source Code: TissueClassification.cxx
XML Description: TissueClassification.xml
Test: TissueClassificationTest.cxx
Usage:
USAGE:
../Slicer3-ext/BrainTissueClassification-build/lib/Slicer3/Plugins/Tissu
eClassification
[--returnparameterfile
<std::string>]
[--processinformationaddress
<std::string>] [--xml] [--echo] [-b
<int>] [-c <int>] [--] [--version]
[-h] <std::string> <std::string>
<std::string> <std::string>
Where:
--returnparameterfile <std::string>
Filename in which to write simple return parameters (int, float,
int-vector, etc.) as opposed to bulk return parameters (image,
geometry, transform, measurement, table).
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
-b <int>, --biasoption <int>
Option for bias correction (0: no bias correction; 1: global linear;
2: global quadratic; 3: region based linear; 4: region based
quadratic) (default: 0)
-c <int>, --class <int>
Number of classes (default: 3)
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
<std::string>
(required) Input T1 Image.
<std::string>
(required) Only voxels inside the mask are classified
<std::string>
(required) Output brain mask map.
<std::string>
(required) Estimated bias field
Description: This module computes voxel by voxel tissue classification
of an MR brain image using a fuzzy c-means algortihm. Bias field is
modeled as a lower order polynomial. Bias field and tissue
classification are estimated iteratively in an EM fashion. Internally,
each voxel is assigned tissue membership function values, which range
from 0 to 1. At any voxel, the sum of membership function of all classes
is either 0 (outside of brain), or 1. The membership functions are
converted in tissue labels to generate hard segmentation.
Author(s): Xiaodong Tao, taox @ research . ge . com
Acknowledgements: This work is part of the National Alliance for Medical
Image Computing (NAMIC), funded by the National Institutes of Health
through the NIH Roadmap for Medical Research, Grant U54 EB005149.
Implementation of the Fuzzy Classification was contributed by Dr.
Ming-Ching Chang from GE Research.
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Implementation of the Fuzzy Classification was contributed by Dr. Ming-Ching Chang from GE Research.
OASIS datasets were used to generate images on this page.
References
- Ming-ching Chang, Xiaodong Tao “Subvoxel Segmentation and Representation of Brain Cortex Using Fuzzy Clustering and Gradient Vector Diffusion”, SPIE Medical Imaging, San Diego, CA, 2010.