Difference between revisions of "Modules:FetchMI-Documentation-3.4"
| Line 32: | Line 32: | ||
== Usage == | == Usage == | ||
| − | === | + | ===Required software & Tutorials=== |
| − | + | In order to use the http://localhost:8081 server provided by default in this module, XNAT Desktop software must be downloaded and installed. The following links provide information on where to go and how to install the software. After software is installed, run XNAT Desktop on your local machine and make sure that webservices are running ( View->Preferences->Server properties: selecte the ''Start server'' button.) | |
| − | * [http:// | + | |
| − | * [http://slicer.spl.harvard.edu/slicerWiki/index.php/Slicer3:XND | + | * [http://nrg.wikispaces.com/XNAT+Desktop+Technical+Specification XNAT Desktop technical specification ] |
| + | * [http://nrg.wustl.edu/xnd/download/ Download XNAT Desktop Version 0.8] | ||
| + | * [http://www.xnat.org/xnd/ XNAT Desktop Installation and Tutorial ] | ||
| + | * [http://slicer.spl.harvard.edu/slicerWiki/index.php/Slicer3:XND Using XNAT Desktop with XNAT Enterprise] | ||
==== Brief instruction==== | ==== Brief instruction==== | ||
Revision as of 18:42, 3 March 2009
Home < Modules:FetchMI-Documentation-3.4Return to Slicer 3.4 Documentation
Module Name
Fetch Medical Informatics (FetchMI)
General Information
Module Type & Category
Type: Interactive
Category: Informatics
Authors, Collaborators & Contact
- Author1: Wendy Plesniak
- Contributor1: Nicole Aucoin
- Contributor2: Steve Pieper
- Contact: Wendy Plesniak, wjp@bwh.harvard.edu
Module Description
FetchMI (Fetch Medical Informatics) is a 'sandbox' functionality being developed to remotely upload, download and tag Slicer scenes and datasets. Its functionality will eventually be moved into Slicer's base code (into the load and save panels) and into a separate module for data markup. Currently, FetchMI interoperates with XNAT Desktop's web services. Its functionality is being extended to work with BIRN's HID and XNAT Enterprise web services.
The module can be used to browse remote repositories for data, download MRML scenes into Slicer, associate metadata with loaded data and the scene description, and upload a scene and its referenced data to a remote host. The interface also permits remote deleting of MRML scenes and their referenced resources.
The module is a new offering within Slicer, and has not yet been widely tested. Please help us to improve it by reporting any unexpected behavior to Slicer's bug tracker.
Usage
Required software & Tutorials
In order to use the http://localhost:8081 server provided by default in this module, XNAT Desktop software must be downloaded and installed. The following links provide information on where to go and how to install the software. After software is installed, run XNAT Desktop on your local machine and make sure that webservices are running ( View->Preferences->Server properties: selecte the Start server button.)
- XNAT Desktop technical specification
- Download XNAT Desktop Version 0.8
- XNAT Desktop Installation and Tutorial
- Using XNAT Desktop with XNAT Enterprise
Brief instruction
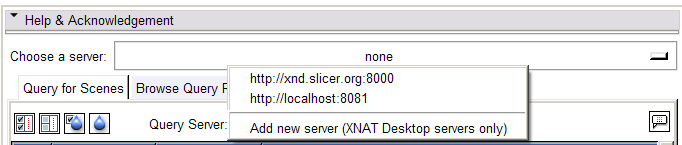
- Select a server. All subsequent operations will transact with the selected server.
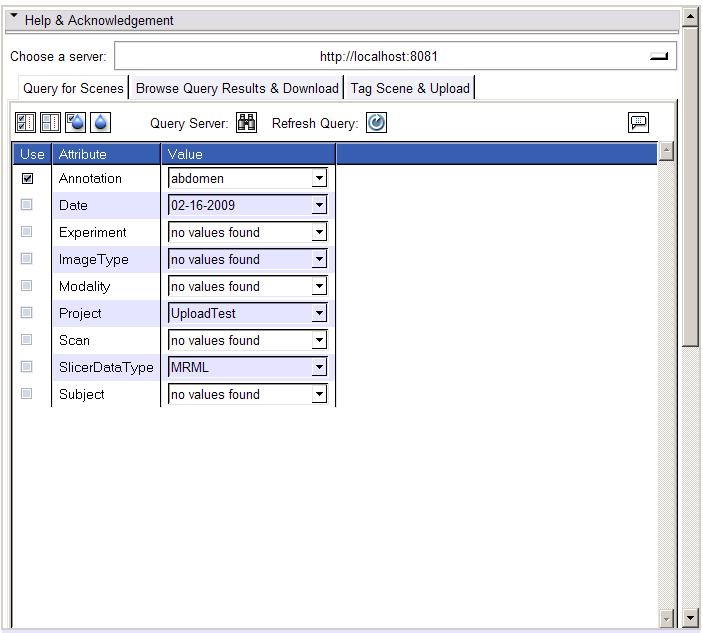
- The selected server will automatically be queried for the metadata it currently supports. The panel in the Query for Scenes tab will be populated with this metadata (attributes and available values). Update this panel time by clicking the Refresh button.
- Select any tags you want to use to query for resources
- Click the Search button (binoculars) to query the server for matching resources.
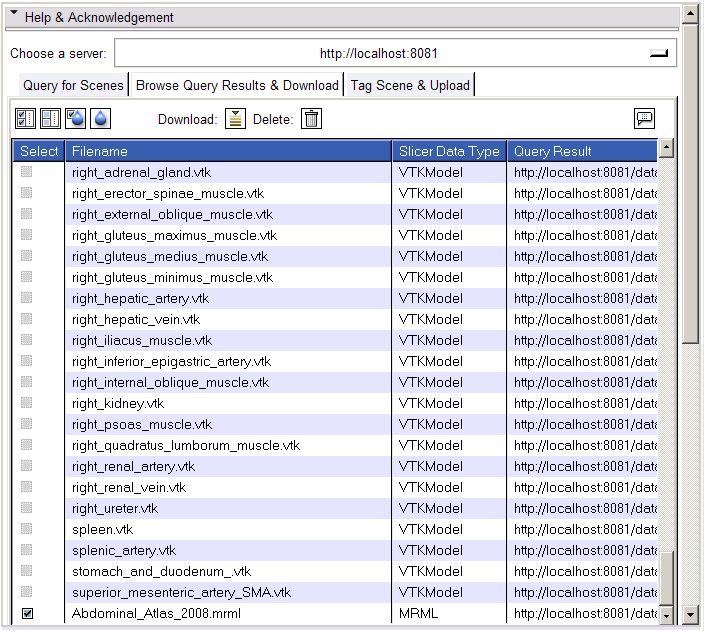
- The Browse Query Results & Download tab will be populated by a list of resources matching the query.
- Select a resource (MRML Scenes only) from that list.
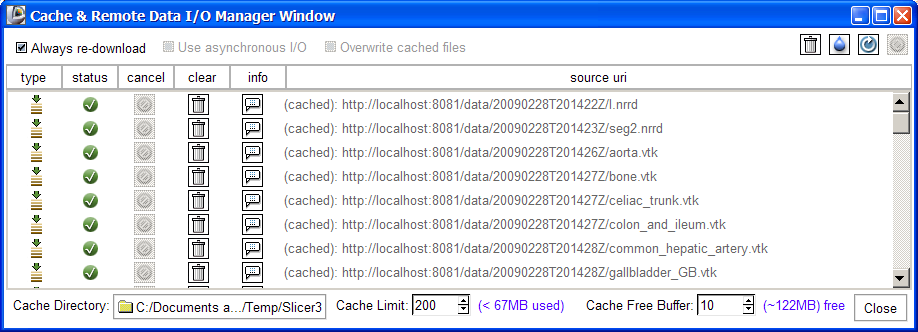
- Click the Download button and the scene will be downloaded to your current cache directory and loaded.
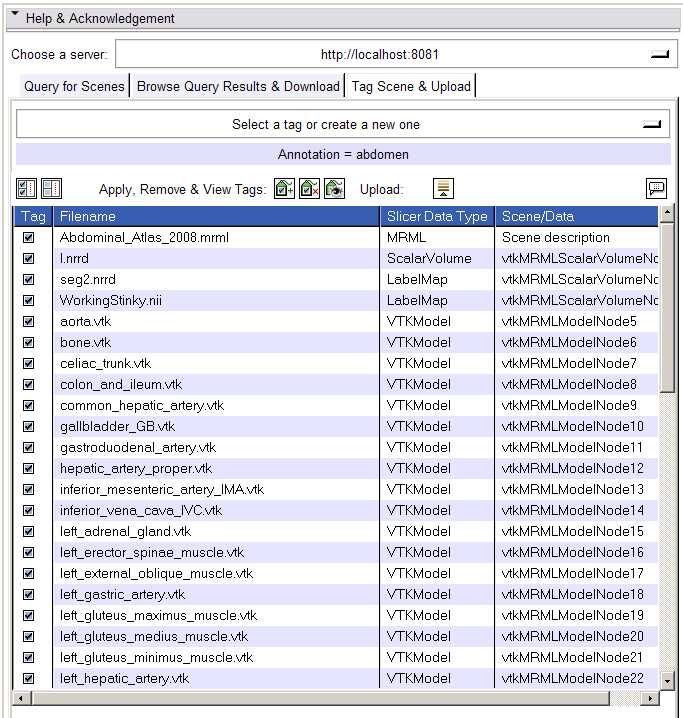
- The Tag Scene & Upload tab shows all data in scene (and scene file).
- Select or create new tags using the menubutton at the top of this panel, and click the Apply Tags button to associate to selected datasets. Click the Remove Tags button to dissociate selected tag from selected datasets.
- NOTE: tags should be brief. At this time, tags CANNOT include SPACES or any special characters; we will admit these options in future development.
- Click the Show All Tags button to see all tags on all loaded datasets and scene.
- Upload the current scene to the server by clicking the Upload button, after data is tagged. In this version, the entire scene and all datasets are uploaded.
- NOTE: remote uploads and downloads can be slow. Though we have attempted to construct safeguards, Initiating these without access to the network or without access to the selected server may still lead to unexpected application behavior. Please report any unexpected behavior to Slicer's bug tracker.
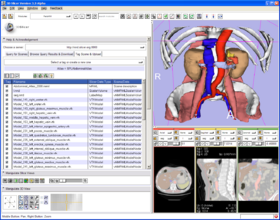
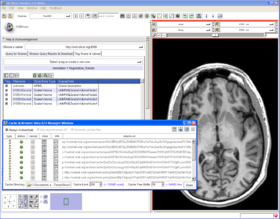
Quick Tour of Features and Use
Below is a tour of interfaces to FetchMI's functionality, with explanatory information and suggestions for use:
- Select a web service:
- Query for Scenes tab:
- Browse Query Results & Download tab:
- Tag Scene & Upload tab:
Development
Dependencies
Other modules or packages that are required for this module's use.
Known bugs
FetchMI is a new and developing module -- please help us by reporting any bugs you encounter. Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Customize following links for your module.
Links to documentation generated by doxygen.
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.