Difference between revisions of "Modules:FastMarchingSegmentation-Documentation-3.5"
| (10 intermediate revisions by one other user not shown) | |||
| Line 5: | Line 5: | ||
{| | {| | ||
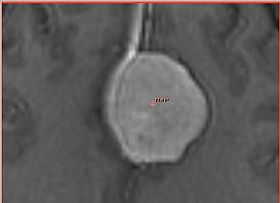
| − | |[[Image:fastmarching-initialization.jpg|thumb|280px|Initialization of the algorithm with fiducial point]] | + | |[[Image:fastmarching-initialization.jpg|thumb|280px|Initialization of the algorithm with a single fiducial point]] |
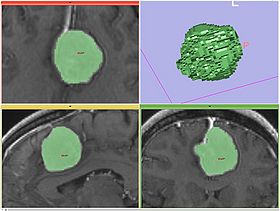
| − | |[[Image:fastmarching-result-adjusted.jpg|thumb|280px|Adjusted segmentation | + | |[[Image:fastmarching-result-adjusted.jpg|thumb|280px|Adjusted segmentation result and volume rendering]] |
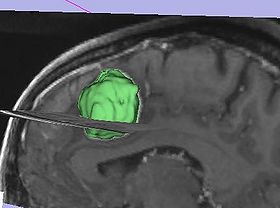
| − | |[[Image:fastmarching- | + | |[[Image:fastmarching-model.jpg|thumb|280px|Model of the resulting tumor segmentation]] |
|} | |} | ||
| Line 35: | Line 35: | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | |||
| − | * '''Input | + | {| |
| − | * ''' | + | | |
| − | * '''Output panel:''' | + | Segmentation workflow of this module consists of the two steps: Initialization and adjustment of the segmentation result. |
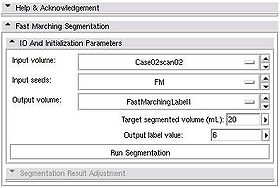
| − | * ''' | + | * '''Initialization:''' The user is required to specify the following items to initialize the segmentation process: |
| + | ** ''Input volume'': image that contains the structure to be segmented | ||
| + | ** ''Input seeds'': fiducial list containing fiducials marking points within the region to be segmented. | ||
| + | ** ''Output volume'': output label volume that will keep the segmentation result | ||
| + | ** ''Target segmented volume'': positive number that estimates expected volume of the segmented structure in milliliters (mL) | ||
| + | ** ''Output label value'': value that will be assigned to the non-zero pixels of the segmentation result | ||
| + | Once these items have been specified, push ''Run Segmentation'' button to initiate the segmentation process. | ||
| + | * '''Adjustment of the segmentation result:''' The segmentation process can be described on the high level as evolution of contours, that starts from the seed points provided in the initialization step. In the current adjustment step, the user can go back in the evolution of the segmentation contour by changing the current value of the volume for the segmented structure. This process is facilitated by the following controls: | ||
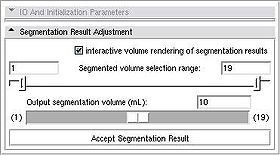
| + | ** ''Segmented volume selection range'': This range slider allows to specify the range of volumes that you want to experiment with in adjusting the result. The maximum range is from 0 to the target segmented volume specified in the previous step. | ||
| + | ** ''Output segmentation volume:'' This slider allows to smoothly scroll through the volume values within the selected range. The segmentation contour is interactively updated in the slice view as the slider is moved. Note, that as you lock in to the most desirable segmentation result, you would reduce the selection range to allow for better precision of contour adjustment. | ||
| + | ** ''Interactive volume rendering of segmentation results'': If this checkbox is selected, the volume rendering of the segmented label will be shown in the 3D View window. This allows to quickly assess the accuracy of segmentation, and identify leaks outside the desired region. | ||
| + | Once the satisfactory result is achieved, ''Accept Segmentation Result'' button will finalize the segmentation. | ||
| + | | | ||
| + | {| | ||
| + | |[[Image:FastMarching-initialization-controls.jpg|thumb|280px|Control panel for FastMarching initialization]] | ||
| + | [[Image:FastMarching-result-controls.jpg|thumb|280px|Control panel for FastMarching result adjustment]] | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | '''Usage tips:''' | ||
| + | * ''Put bigger number as the estimate volume'' It is better to overestimate the volume of the structure in the initialization, than underestimate. Remember, that you can always scroll back in the contour evolution, but cannot exceed predefined volume limit. | ||
| + | * ''Play with the fiducial points'' If the segmentation result is not satisfactory, try adding more fiducials. If the region to be segmented has regions of different intensities, put fiducials in each of such regions. If all fails, try to segment separate regions one after another, and use [[Modules:ImageLabelCombine-3.4|ImageLabelCombine]] module to merge them together later. | ||
| + | * ''Take advantage of volume rendering'' You can quickly see if there are "leaks", without the need to scroll through the slices. However, volume rendering may not show small "leaks". | ||
== Development == | == Development == | ||
| − | + | Some of the desired features: | |
| − | |||
| − | |||
| − | |||
| − | |||
* add ability to initialize with label, not only with fiducials | * add ability to initialize with label, not only with fiducials | ||
* documentation | * documentation | ||
| − | |||
* integration with Editor | * integration with Editor | ||
| − | |||
* add parameter node | * add parameter node | ||
* Ron: specify the target segmented volume in either mm^3 or cm^3 or major dimensions (1x2x4cm) | * Ron: specify the target segmented volume in either mm^3 or cm^3 or major dimensions (1x2x4cm) | ||
| − | |||
===Dependencies=== | ===Dependencies=== | ||
| − | + | Core modules: [[Modules:Fiducials-Documentation-3.4|Fiducials]], [[Modules:Volumes-Documentation-3.4|Volumes]] | |
===Known bugs=== | ===Known bugs=== | ||
Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. | ||
| − | |||
| − | |||
===Usability issues=== | ===Usability issues=== | ||
| Line 76: | Line 88: | ||
Source code can accessed [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/FastMarchingSegmentation/ here] | Source code can accessed [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/FastMarchingSegmentation/ here] | ||
| − | [http://www.na-mic.org/Slicer/Documentation/Slicer3/html/ Links] to documentation generated by doxygen. | + | [http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/ Links] to documentation generated by doxygen. |
== More Information == | == More Information == | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
| − | Supported by | + | {| |
| + | |[[Image:fastmarching-acks.jpg|thumb|280px|Supported by BrainScienceFoundation]] | ||
| + | |} | ||
===References=== | ===References=== | ||
* Pichon E, Tannenbaum A, Kikinis R. A statistically based flow for image segmentation. Med Image Anal. 2004 Sep;8(3):267-74. PMID: 15450221. [http://www.spl.harvard.edu/publications/item/view/193] | * Pichon E, Tannenbaum A, Kikinis R. A statistically based flow for image segmentation. Med Image Anal. 2004 Sep;8(3):267-74. PMID: 15450221. [http://www.spl.harvard.edu/publications/item/view/193] | ||
Latest revision as of 23:05, 15 January 2010
Home < Modules:FastMarchingSegmentation-Documentation-3.5Return to Slicer 3.5 Documentation
Fast Marching Segmentation
FastMarchingSegmentation
General Information
Module Type & Category
Type: Interactive
Category: Segmentation
Authors, Collaborators & Contact
- Andriy Fedorov, BWH
- Eric Pichon
- Contact: Andriy Fedorov, fedorov at bwh
Module Description
This module implements segmentation based on fast marching algorithm.
Usage
Examples, Use Cases & Tutorials
- This module is most useful to segment image regions that have similar intensity
- Initialization is very simple: points within the region to be segmented and expected volume of the segmented structure
- The segmentation is completed relatively quickly for typical images, facilitating experimentation with the selection of optimum parameters
- The resulting volume can be adjusted interactively by scrolling through the evolution of the label contour
Quick Tour of Features and Use
|
Segmentation workflow of this module consists of the two steps: Initialization and adjustment of the segmentation result.
Once these items have been specified, push Run Segmentation button to initiate the segmentation process.
Once the satisfactory result is achieved, Accept Segmentation Result button will finalize the segmentation. |
|
Usage tips:
- Put bigger number as the estimate volume It is better to overestimate the volume of the structure in the initialization, than underestimate. Remember, that you can always scroll back in the contour evolution, but cannot exceed predefined volume limit.
- Play with the fiducial points If the segmentation result is not satisfactory, try adding more fiducials. If the region to be segmented has regions of different intensities, put fiducials in each of such regions. If all fails, try to segment separate regions one after another, and use ImageLabelCombine module to merge them together later.
- Take advantage of volume rendering You can quickly see if there are "leaks", without the need to scroll through the slices. However, volume rendering may not show small "leaks".
Development
Some of the desired features:
- add ability to initialize with label, not only with fiducials
- documentation
- integration with Editor
- add parameter node
- Ron: specify the target segmented volume in either mm^3 or cm^3 or major dimensions (1x2x4cm)
Dependencies
Core modules: Fiducials, Volumes
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source code can accessed here
Links to documentation generated by doxygen.
More Information
Acknowledgment
References
- Pichon E, Tannenbaum A, Kikinis R. A statistically based flow for image segmentation. Med Image Anal. 2004 Sep;8(3):267-74. PMID: 15450221. [1]