Difference between revisions of "Modules:EMSegmentBatch-Documentation-3.4"
(Add Register Images BatchMake Module documentation) |
m (Text replacement - "\[http:\/\/wiki\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) ([^]]+)]" to "$2") |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
[[Documentation-3.4|Return to Slicer 3.4 Documentation]] | [[Documentation-3.4|Return to Slicer 3.4 Documentation]] | ||
| + | |||
| + | [[Announcements:Slicer3.4#Highlights|Gallery of New Features]] | ||
__NOTOC__ | __NOTOC__ | ||
===Module Name=== | ===Module Name=== | ||
| − | + | EM Segment BatchMake | |
| + | {| | ||
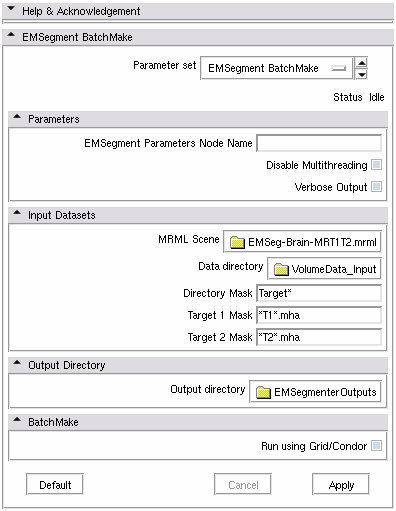
| + | |[[Image:EMSegmentBatchMakeModule.png|thumb|580px|EM Segment BatchMake module GUI ]] | ||
| + | |} | ||
== General Information == | == General Information == | ||
| Line 12: | Line 17: | ||
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | * Authors: Julien Finet | + | * Authors: Stephen Aylward, Julien Finet |
| − | * Contact: | + | * Contact: stephen.aylward at kitware.com |
===Module Description=== | ===Module Description=== | ||
| − | This | + | This module is the BatchMake version of [[Modules:EMSegment-Simple-Documentation-3.4|EM Segment Simple]], a simplified "one-click" GUI interface for the EMSegment Command-line Executable. The interface is simple because the number of required command-line parameters is minimized, and it is flexible because any EM algorithm parameter can be modified, within the MRML scene, via the EMSegment GUI interface. |
| + | The batch processing feature allows the user to run the EMSegment module on multiple patients. By defining a patient root directory and a mask to iterate through the patients, EMSegment simple will be run for each images found in the matched images. | ||
== Usage == | == Usage == | ||
| − | + | MRML Scene = the mrml file name that contains all the parameters previously defined with EMSegment Template Builder module<br> | |
| − | + | Data directory = where the patients images are ( T1, T2 images ) | |
| − | + | Directory mask = to be a valid input, the patients directories located in "Data directory" must match the mask | |
| − | + | Target 1 mask = T1 image mask that must be matched inside the patients directories | |
| − | + | Target 2 mask = T2 image mask that must be matched inside the patients directories | |
| − | + | Output directory = where the results are saved | |
| − | |||
===Examples, Use Cases & Tutorials=== | ===Examples, Use Cases & Tutorials=== | ||
| Line 31: | Line 36: | ||
For the following file tree:<br> | For the following file tree:<br> | ||
* Images | * Images | ||
| − | ** | + | ** EMSegmentParameters.mrml |
| + | ** Atlas | ||
| + | *** ... | ||
** Patient1 | ** Patient1 | ||
| − | *** | + | *** Patient1.txt |
| − | *** | + | *** Patient1-T1.mha |
| + | *** Patient1-T2.mha | ||
** Patient2 | ** Patient2 | ||
| − | *** | + | *** Patient2.txt |
| − | *** | + | *** Patient2-T1.mha |
| + | *** Patient2-T2.mha | ||
** Patient3 | ** Patient3 | ||
| − | *** | + | *** Patient3.txt |
| − | *** | + | *** Patient3-T1.mha |
| − | * | + | *** Patient3-T2.mha |
| − | Parameters | + | * Results |
| − | + | Parameters are:<br> | |
| − | + | MRMLScene: "Images/EMSegmentParameters.mrml"<br> | |
| − | + | Data Directory: "Images"<br> | |
| − | + | Directory Mask: Patient*<br> | |
| − | + | Target 1 Mask: *T1.mha<br> | |
| + | Target 2 Mask: *T2.mha<br> | ||
| + | Output directory: "Results"<br> | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | To run the module on a condor grid, open the BatchMake parameter group and choose "condor" as running mode. | + | To run the module on a condor grid, open the BatchMake parameter group and choose "condor" as running mode. Make sure your data are accessible by all the condor machines. |
| − | |||
| − | |||
| − | |||
| − | |||
== Development == | == Development == | ||
| Line 61: | Line 68: | ||
===Dependencies=== | ===Dependencies=== | ||
| − | + | EMSegment Simple module | |
===Known bugs=== | ===Known bugs=== | ||
| Line 73: | Line 80: | ||
===Source code & documentation=== | ===Source code & documentation=== | ||
| − | Source Code: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/BatchMakeApplications/ | + | Source Code: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/BatchMakeApplications/EMSegmentBatchMakeModule/EMSegmentBatchMakeModule.cxx?view=annotate EMSegmentBatchMakeModule.cxx] |
| − | XML Description: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/BatchMakeApplications/ | + | XML Description: [http://www.na-mic.org/ViewVC/index.cgi/trunk/Applications/CLI/BatchMakeApplications/EMSegmentBatchMakeModule/EMSegmentBatchMakeModule.xml?view=co EMSegmentBatchMakeModule.xml] |
Documentation: | Documentation: | ||
Latest revision as of 02:16, 27 November 2019
Home < Modules:EMSegmentBatch-Documentation-3.4Return to Slicer 3.4 Documentation
Module Name
EM Segment BatchMake
General Information
Module Type & Category
Type: CLI
Category: Batch Processing
Authors, Collaborators & Contact
- Authors: Stephen Aylward, Julien Finet
- Contact: stephen.aylward at kitware.com
Module Description
This module is the BatchMake version of EM Segment Simple, a simplified "one-click" GUI interface for the EMSegment Command-line Executable. The interface is simple because the number of required command-line parameters is minimized, and it is flexible because any EM algorithm parameter can be modified, within the MRML scene, via the EMSegment GUI interface. The batch processing feature allows the user to run the EMSegment module on multiple patients. By defining a patient root directory and a mask to iterate through the patients, EMSegment simple will be run for each images found in the matched images.
Usage
MRML Scene = the mrml file name that contains all the parameters previously defined with EMSegment Template Builder module
Data directory = where the patients images are ( T1, T2 images )
Directory mask = to be a valid input, the patients directories located in "Data directory" must match the mask
Target 1 mask = T1 image mask that must be matched inside the patients directories
Target 2 mask = T2 image mask that must be matched inside the patients directories
Output directory = where the results are saved
Examples, Use Cases & Tutorials
For the following file tree:
- Images
- EMSegmentParameters.mrml
- Atlas
- ...
- Patient1
- Patient1.txt
- Patient1-T1.mha
- Patient1-T2.mha
- Patient2
- Patient2.txt
- Patient2-T1.mha
- Patient2-T2.mha
- Patient3
- Patient3.txt
- Patient3-T1.mha
- Patient3-T2.mha
- Results
Parameters are:
MRMLScene: "Images/EMSegmentParameters.mrml"
Data Directory: "Images"
Directory Mask: Patient*
Target 1 Mask: *T1.mha
Target 2 Mask: *T2.mha
Output directory: "Results"
Quick Tour of Features and Use
To run the module on a condor grid, open the BatchMake parameter group and choose "condor" as running mode. Make sure your data are accessible by all the condor machines.
Development
Dependencies
EMSegment Simple module
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source Code: EMSegmentBatchMakeModule.cxx
XML Description: EMSegmentBatchMakeModule.xml
Documentation:
More Information
Acknowledgment
This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on the National Centers for Biomedical Computing can be obtained from National Centers for Biomedical Computing.