Difference between revisions of "Modules:EMSegment-TemplateBuilder:EMSegment-TemplateBuilder-Steps"
From Slicer Wiki
(New page: = EMSegment Template Builder Workflow Wizard Steps= == (1/9)Define Parameters Set: Select parameter set or create new parameters == [[Image:EMSegment-Workflow-1-9.png|thumb|580px|Step 1/9...) |
|||
| Line 1: | Line 1: | ||
= EMSegment Template Builder Workflow Wizard Steps= | = EMSegment Template Builder Workflow Wizard Steps= | ||
| − | == (1/9)Define Parameters Set: Select parameter set or create new parameters == | + | == (1/9) Define Parameters Set: Select parameter set or create new parameters == |
[[Image:EMSegment-Workflow-1-9.png|thumb|580px|Step 1/9]] | [[Image:EMSegment-Workflow-1-9.png|thumb|580px|Step 1/9]] | ||
* For now, use tutorial set | * For now, use tutorial set | ||
| Line 7: | Line 7: | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | == (2/9)Define Hierarchy: Define a hierarchy of anatomical structures == | + | == (2/9) Define Hierarchy: Define a hierarchy of anatomical structures == |
[[Image:EMSegment-Workflow-2-9.png|thumb|580px|Step 2/9]] | [[Image:EMSegment-Workflow-2-9.png|thumb|580px|Step 2/9]] | ||
* Right click to add or delete nodes | * Right click to add or delete nodes | ||
| Line 13: | Line 13: | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | == (3/9)Assign Atlas: Assign atlases for anatomical structures == | + | == (3/9) Assign Atlas: Assign atlases for anatomical structures == |
[[Image:EMSegment-Workflow-3-9.png|thumb|580px|Step 3/9]] | [[Image:EMSegment-Workflow-3-9.png|thumb|580px|Step 3/9]] | ||
| + | * To change or add new atlas volumes: Load new volumes into Slicer3, then Select new volumes at this step | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | == (4/9)Select Target Images: Choose the set of images that will be segmented == | + | == (4/9) Select Target Images: Choose the set of images that will be segmented == |
| + | * To change or add new atlas volumes: Load new volumes into Slicer3, then Select new volumes at this step | ||
| + | * You can reorder target images; order is important | ||
| + | * You can choose to align target images | ||
| + | * First target is fixed image | ||
| + | * Rigid, mutual information registration | ||
[[Image:EMSegment-Workflow-4-9.png|thumb|580px|Step 4/9]] | [[Image:EMSegment-Workflow-4-9.png|thumb|580px|Step 4/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | == (5/9)Intensity Normalization: Normalize target images == | + | == (5/9) Intensity Normalization: Normalize target images == |
| + | * You can choose to normalize target images | ||
| + | * Simple, default strategy | ||
| + | * Default parameter sets available from pulldown | ||
[[Image:EMSegment-Workflow-5-9.png|thumb|580px|Step 5/9]] | [[Image:EMSegment-Workflow-5-9.png|thumb|580px|Step 5/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
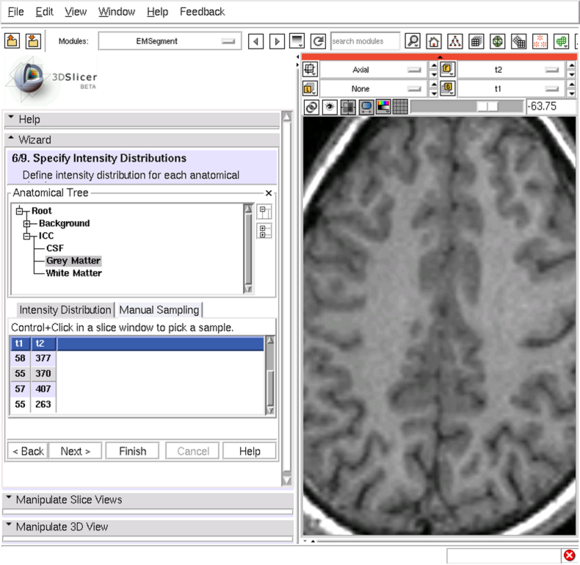
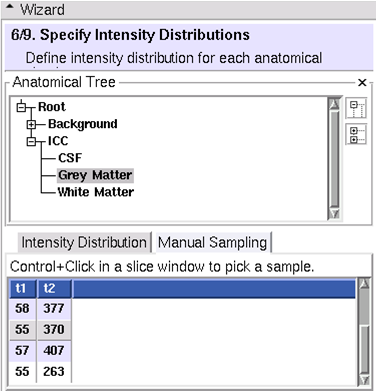
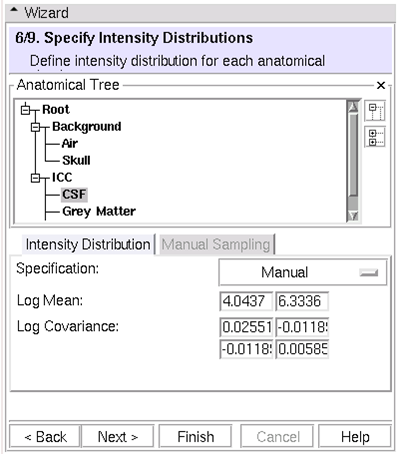
| − | == (6/9)Specify Intensity Distributions: Define intensity distribution for each anatomical structure == | + | == (6/9) Specify Intensity Distributions: Define intensity distribution for each anatomical structure == |
| + | * Intensity distributions define appearance of each leaf structure | ||
| + | * Gaussian | ||
| + | * Dimensionality equal to number of target images | ||
| + | * Two methods | ||
| + | ** Sample voxels from images | ||
| + | [[Image:EMSegment-Workflow-6-9-sample.png|thumb|580px|Step 6/9 - Sample Voxels]] [[Image:EMSegment-Workflow-6-9-sample-remove.png|thumb|580px|Step 6/9 - Remove Voxels]] | ||
| + | *** Load first target image into Slicer3 slicer view | ||
| + | *** Choose anatomical structure | ||
| + | *** Choose `manual sampling’ | ||
| + | *** Ctrl-left-click on image to add voxels | ||
| + | *** To remove an unwanted sample right-click on it and choose “remove” | ||
| + | *** Change back to “manual” mode to tweak distribution | ||
| + | ** Specify mean and covariance manually | ||
| + | * Tip: sample first, then fine tune manually | ||
| + | |||
[[Image:EMSegment-Workflow-6-9.png|thumb|580px|Step 6/9]] | [[Image:EMSegment-Workflow-6-9.png|thumb|580px|Step 6/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
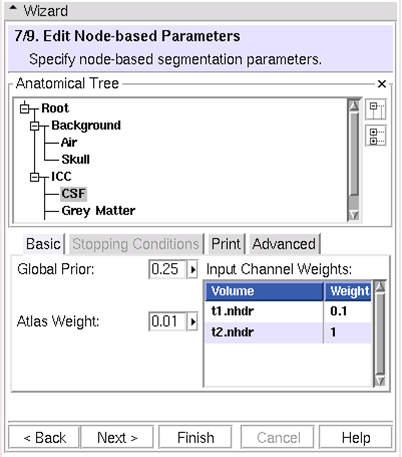
| − | == (7/9)Edit Node-based Parameters: Specify node-based segmentation parameters == | + | == (7/9) Edit Node-based Parameters: Specify node-based segmentation parameters == |
| + | * Segmentation parameters for every tree node | ||
| + | * Influence | ||
| + | ** Prior weight relative to other structures | ||
| + | ** Atlas | ||
| + | ** Input channels | ||
| + | ** Smoothing (parent nodes only) | ||
| + | * Stopping Conditions | ||
[[Image:EMSegment-Workflow-7-9.png|thumb|580px|Step 7/9]] | [[Image:EMSegment-Workflow-7-9.png|thumb|580px|Step 7/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
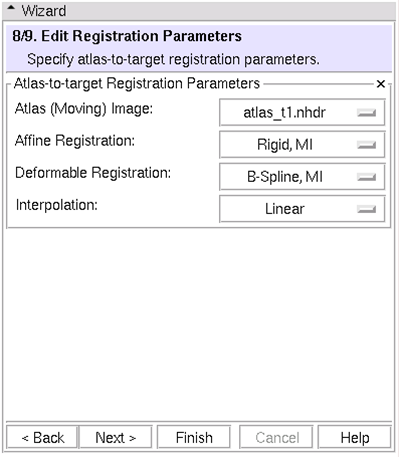
| − | == (8/9)Edit Registration Parameters: Specify atlas-to-target registration parameters == | + | == (8/9) Edit Registration Parameters: Specify atlas-to-target registration parameters == |
| + | * Moving image registered to first target image (You can choose any image loaded into Slicer3) | ||
| + | * Same transformation applied to all atlas images before segmentation begins | ||
[[Image:EMSegment-Workflow-8-9.png|thumb|580px|Step 8/9]] | [[Image:EMSegment-Workflow-8-9.png|thumb|580px|Step 8/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| − | == (9/9)Run Segmentation: Save work and apply EM Algorithm to segment target images == | + | ==(9/9) Run Segmentation: Save work and apply EM Algorithm to segment target images== |
| + | * Select output and run registration | ||
| + | * You can troubleshoot preprocessing by saving intermediate results | ||
| + | * Choose an output labelmap | ||
| + | * ROI governs segmentation processing (one-based, not zero-based) | ||
| + | |||
[[Image:EMSegment-Workflow-9-9.png|thumb|580px|Step 9/9]] | [[Image:EMSegment-Workflow-9-9.png|thumb|580px|Step 9/9]] | ||
<br style="clear:both;"/> | <br style="clear:both;"/> | ||
| Line 43: | Line 81: | ||
= Generate EM Segmentation Result Gallery = | = Generate EM Segmentation Result Gallery = | ||
| − | + | * Select the first target volume for display | |
| − | + | * Select the segmentation results volume in the Labelmap chooser | |
| − | + | * Adjust opacity of the label map | |
| − | + | * (Optional) Generate and display surfaces from the segmentation results | |
| − | + | # Select the "Model Generation" -> "Model Maker" module | |
| − | + | # For "Input Volume", select the $My_Segmentation_Result | |
| − | + | # For "Output Directory", create and select $PLAYPEN_DIR/Models | |
| − | + | # For "Model Scene File", select $PLAYPEN_DIR/Models.mrml | |
| − | + | # Enable "Generate All Models" | |
| − | + | # Set "End Label" to the highest label that you produced (e.g., 8 fro the tutorial) | |
| − | + | # Enable "Joint Smoothing" | |
| − | + | # Click on Apply (this will generate the models and will take approximately 5 minutes) | |
| − | + | # Load the models into Slicer (File->Import Scene; select $PLAYPEN_DIR/Models.mrml) | |
| − | + | # Use The Models module to manipulate display attributes | |
| − | |||
Revision as of 17:48, 19 March 2009
Home < Modules:EMSegment-TemplateBuilder:EMSegment-TemplateBuilder-StepsContents
- 1 EMSegment Template Builder Workflow Wizard Steps
- 1.1 (1/9) Define Parameters Set: Select parameter set or create new parameters
- 1.2 (2/9) Define Hierarchy: Define a hierarchy of anatomical structures
- 1.3 (3/9) Assign Atlas: Assign atlases for anatomical structures
- 1.4 (4/9) Select Target Images: Choose the set of images that will be segmented
- 1.5 (5/9) Intensity Normalization: Normalize target images
- 1.6 (6/9) Specify Intensity Distributions: Define intensity distribution for each anatomical structure
- 1.7 (7/9) Edit Node-based Parameters: Specify node-based segmentation parameters
- 1.8 (8/9) Edit Registration Parameters: Specify atlas-to-target registration parameters
- 1.9 (9/9) Run Segmentation: Save work and apply EM Algorithm to segment target images
- 2 Generate EM Segmentation Result Gallery
EMSegment Template Builder Workflow Wizard Steps
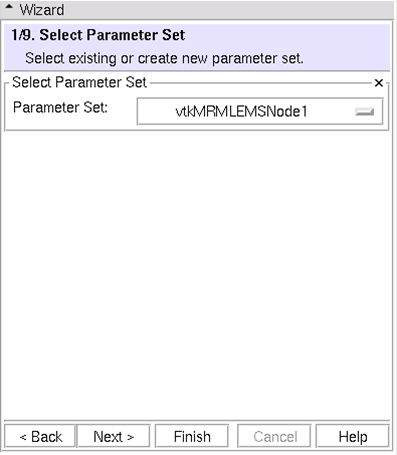
(1/9) Define Parameters Set: Select parameter set or create new parameters
* For now, use tutorial set * Later use this interface to create a new parameter set
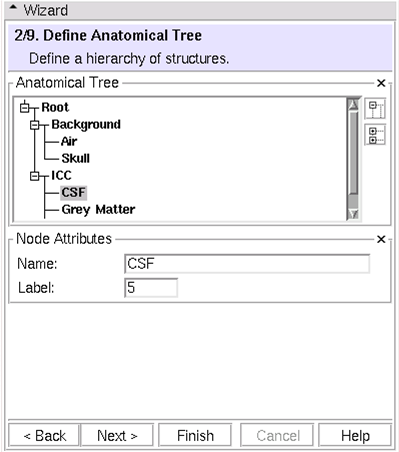
(2/9) Define Hierarchy: Define a hierarchy of anatomical structures
* Right click to add or delete nodes * Label corresponds to eventual voxel values in segmentation result
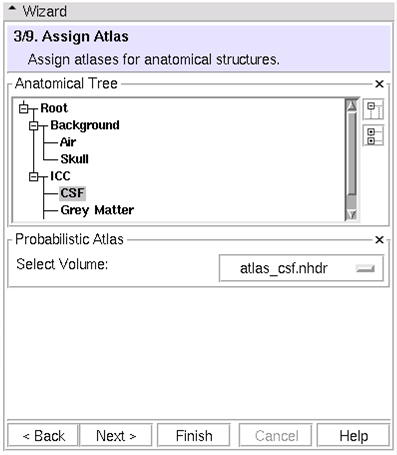
(3/9) Assign Atlas: Assign atlases for anatomical structures
- To change or add new atlas volumes: Load new volumes into Slicer3, then Select new volumes at this step
(4/9) Select Target Images: Choose the set of images that will be segmented
- To change or add new atlas volumes: Load new volumes into Slicer3, then Select new volumes at this step
- You can reorder target images; order is important
- You can choose to align target images
- First target is fixed image
- Rigid, mutual information registration
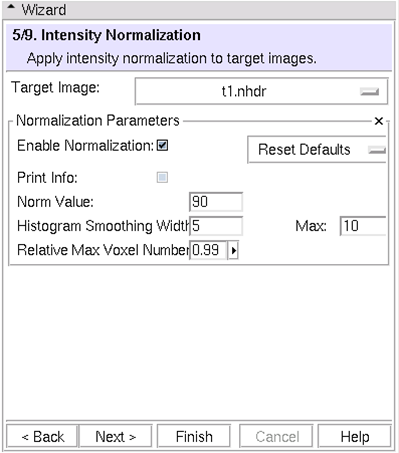
(5/9) Intensity Normalization: Normalize target images
- You can choose to normalize target images
- Simple, default strategy
- Default parameter sets available from pulldown
(6/9) Specify Intensity Distributions: Define intensity distribution for each anatomical structure
- Intensity distributions define appearance of each leaf structure
- Gaussian
- Dimensionality equal to number of target images
- Two methods
- Sample voxels from images
- Load first target image into Slicer3 slicer view
- Choose anatomical structure
- Choose `manual sampling’
- Ctrl-left-click on image to add voxels
- To remove an unwanted sample right-click on it and choose “remove”
- Change back to “manual” mode to tweak distribution
- Specify mean and covariance manually
- Tip: sample first, then fine tune manually
(7/9) Edit Node-based Parameters: Specify node-based segmentation parameters
- Segmentation parameters for every tree node
- Influence
- Prior weight relative to other structures
- Atlas
- Input channels
- Smoothing (parent nodes only)
- Stopping Conditions
(8/9) Edit Registration Parameters: Specify atlas-to-target registration parameters
- Moving image registered to first target image (You can choose any image loaded into Slicer3)
- Same transformation applied to all atlas images before segmentation begins
(9/9) Run Segmentation: Save work and apply EM Algorithm to segment target images
- Select output and run registration
- You can troubleshoot preprocessing by saving intermediate results
- Choose an output labelmap
- ROI governs segmentation processing (one-based, not zero-based)
Generate EM Segmentation Result Gallery
- Select the first target volume for display
- Select the segmentation results volume in the Labelmap chooser
- Adjust opacity of the label map
- (Optional) Generate and display surfaces from the segmentation results
- Select the "Model Generation" -> "Model Maker" module
- For "Input Volume", select the $My_Segmentation_Result
- For "Output Directory", create and select $PLAYPEN_DIR/Models
- For "Model Scene File", select $PLAYPEN_DIR/Models.mrml
- Enable "Generate All Models"
- Set "End Label" to the highest label that you produced (e.g., 8 fro the tutorial)
- Enable "Joint Smoothing"
- Click on Apply (this will generate the models and will take approximately 5 minutes)
- Load the models into Slicer (File->Import Scene; select $PLAYPEN_DIR/Models.mrml)
- Use The Models module to manipulate display attributes