Difference between revisions of "Modules:EMSegment-Simple-3.4"
Belhachemi (talk | contribs) m (moved Modules:EMSegment-Simple to Modules:EMSegment-Simple-3.4: Refactoring the EM Wiki Pages) |
|||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
[[Documentation-3.4|Return to Slicer 3.4 Documentation]] | [[Documentation-3.4|Return to Slicer 3.4 Documentation]] | ||
| + | |||
| + | [[Announcements:Slicer3.4#Highlights|Gallery of New Features]] | ||
__NOTOC__ | __NOTOC__ | ||
===Module Name=== | ===Module Name=== | ||
EM Segment Simple | EM Segment Simple | ||
{| | {| | ||
| − | |||
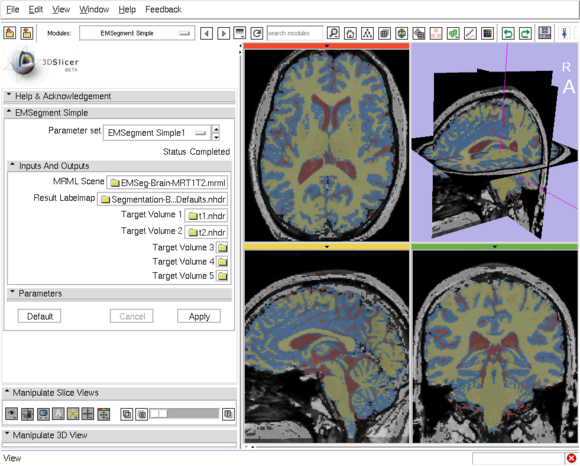
|[[Image:EMSegment-Simple.png|thumb|580px|EM Segment Simple module GUI and Result]] | |[[Image:EMSegment-Simple.png|thumb|580px|EM Segment Simple module GUI and Result]] | ||
|} | |} | ||
| Line 16: | Line 17: | ||
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | |||
* Brad Davis: Kitware, Inc. | * Brad Davis: Kitware, Inc. | ||
| + | * Kilian Pohl: IBM | ||
* Sebastien Barre: Kitware, Inc. | * Sebastien Barre: Kitware, Inc. | ||
* Yumin Yuan: Kitware, Inc. | * Yumin Yuan: Kitware, Inc. | ||
| Line 30: | Line 31: | ||
===Examples, Use Cases & Tutorials=== | ===Examples, Use Cases & Tutorials=== | ||
| + | * A full Tutorial for all three EMSegment modules | ||
| + | |||
| + | These slides and data describe the Slicer3 implementation of all three EMSegment modules and demonstrate their use. A recent (later than 7Jan08) version of Slicer3 is required to run the test data. | ||
| − | + | [[Media:EMSegTutorial-AHM2008.ppt | Slides]] - [[Media:EMSegTutorial-AHM2008.zip | Data]] | |
| − | |||
| − | |||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| − | |||
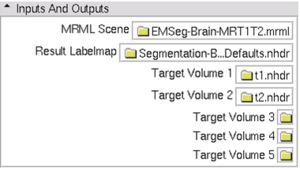
| − | * ''' | + | * '''Parameters panel:''' [[Image:EMSegment-Simple-Panel.png|300px|thumb|EMSegment Simple Parameters Panel]] |
| − | * | + | ** MRML Scene: Select the mrml scene for the tutorial parameter set |
| − | * | + | ** Result Labelmap: Select a new image filename for output labelmap |
| − | * | + | ** Target Volumes: Select input target volumes (Note: target volume order is important) |
| + | ** Click `Apply’, wait (~30 minutes for full size images) | ||
== Development == | == Development == | ||
| − | + | The module is implemented as a very simple programmatic Slicer3 module, whose purpose is to offer simple interface for segmenting new data | |
| − | + | with predefined parameter set. The module GUI interacts with the Logic classes, which are solely responsible for maintaining and accessing MRML nodes. The | |
| − | The module is implemented as a programmatic Slicer3 module | + | Logic class provides APIs that the GUI code uses to access and modify data. The Logic class also wraps the algorithm code itself. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | Logic class provides | ||
| − | modify data. The Logic class also wraps the algorithm code itself. | ||
===Dependencies=== | ===Dependencies=== | ||
| Line 69: | Line 64: | ||
Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. Please select the '''usability issue category''' when browsing or contributing. | Follow this [http://na-mic.org/Mantis/main_page.php link] to the Slicer3 bug tracker. Please select the '''usability issue category''' when browsing or contributing. | ||
| − | |||
| − | |||
===Source code & documentation=== | ===Source code & documentation=== | ||
| Line 76: | Line 69: | ||
[http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/EMSegment/ Links] for the module. | [http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/EMSegment/ Links] for the module. | ||
*[http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/EMSegment/ View EMSegmenter Code] | *[http://www.na-mic.org/ViewVC/index.cgi/trunk/Modules/EMSegment/ View EMSegmenter Code] | ||
| − | *API: [http://www.na-mic.org/Slicer/Documentation/Slicer3/html/classvtkEMSegmentMRMLManager.html MRML Manager] | [http://www.na-mic.org/Slicer/Documentation/Slicer3/html/classvtkEMSegmentLogic.html Module Logic] | [http://www.na-mic.org/Slicer/Documentation/Slicer3/html/classvtkEMSegmentGUI.html Module GUI] | + | *API: [http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/classvtkEMSegmentMRMLManager.html MRML Manager] | [http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/classvtkEMSegmentLogic.html Module Logic] | [http://www.na-mic.org/Slicer/Documentation/Slicer3-doc/html/classvtkEMSegmentGUI.html Module GUI] |
| − | |||
== More Information == | == More Information == | ||
For more information about the EM Segmenter project in Slicer3 [[Slicer3:EM|see the old EMSegment Wiki page here]] | For more information about the EM Segmenter project in Slicer3 [[Slicer3:EM|see the old EMSegment Wiki page here]] | ||
| + | |||
| + | The old [[Modules:EM-Segmenter-Documentation:FAQ |EMSegmenter FAQ]] contains frequently asked questions about the EMSegment interfaces in Slicer3. | ||
| + | |||
| + | There is also an old [http://www.na-mic.org/Wiki/index.php/EMSegmenter_Mini-Tutorials EMSegmenter Mini-Tutorials] | ||
===Acknowledgment=== | ===Acknowledgment=== | ||
Latest revision as of 15:08, 19 August 2010
Home < Modules:EMSegment-Simple-3.4Return to Slicer 3.4 Documentation
Module Name
EM Segment Simple
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Brad Davis: Kitware, Inc.
- Kilian Pohl: IBM
- Sebastien Barre: Kitware, Inc.
- Yumin Yuan: Kitware, Inc.
- Polina Golland: MIT
- Contact: Brad Davis, brad.davis@kitware.com
Module Description
This module provide a simplified "one-click" GUI interface for the EMSegment Command-line Executable. The interface is simple because the number of required command-line parameters is minimized, and it is flexible because any EM algorithm parameter can be modified, within the MRML scene, via the EMSegment GUI interface.
Usage
Similar to the EMSegment Command-line Executable, once a set of parameters for segmentation algorithm is deemed satisfactory through the process of calibrating in the Template Builder module, the user will commonly want to bypass this detailed calibration process when segmenting new images. This module provides a simple interface to specify a small number of required command-line parameters --- a MRML scene (containing algorithm parameters), a target image (or multiple target images, e.g., T1 and T2), and an output labelmap image --- , and to allow any EM algorithm parameters to be modified, within the MRML scene, via the EMSegment GUI interface.
Examples, Use Cases & Tutorials
- A full Tutorial for all three EMSegment modules
These slides and data describe the Slicer3 implementation of all three EMSegment modules and demonstrate their use. A recent (later than 7Jan08) version of Slicer3 is required to run the test data.
Quick Tour of Features and Use
- Parameters panel:
- MRML Scene: Select the mrml scene for the tutorial parameter set
- Result Labelmap: Select a new image filename for output labelmap
- Target Volumes: Select input target volumes (Note: target volume order is important)
- Click `Apply’, wait (~30 minutes for full size images)
Development
The module is implemented as a very simple programmatic Slicer3 module, whose purpose is to offer simple interface for segmenting new data with predefined parameter set. The module GUI interacts with the Logic classes, which are solely responsible for maintaining and accessing MRML nodes. The Logic class provides APIs that the GUI code uses to access and modify data. The Logic class also wraps the algorithm code itself.
Dependencies
Slicer3 base modules.
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
The EMSegmenter can be adapted to many segmentation problems. However, there is no "default" set of parameters that will work for all segmentation problems.
- Atlas-to-target registration and intensity normalization are very important; it will be most effective to apply these steps using algorithms that are customized to your data. Defaults are provided but they may perform poorly for your data---this will lead to poor segmentation results.
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links for the module.
More Information
For more information about the EM Segmenter project in Slicer3 see the old EMSegment Wiki page here
The old EMSegmenter FAQ contains frequently asked questions about the EMSegment interfaces in Slicer3.
There is also an old EMSegmenter Mini-Tutorials
Acknowledgment
Funding for the EMSegmenter module was provided by NAMIC.
References
Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212. [bib]