Difference between revisions of "Modules:EMSegment-Command-Line3.6"
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
EM Segment Command-Line Executable | EM Segment Command-Line Executable | ||
{| | {| | ||
| − | |[[Image:EMSegment-CL-Panel.png|thumb| | + | |[[Image:EMSegment-CL-Panel.png|thumb|400px|Interface]] |
|} | |} | ||
| Line 42: | Line 42: | ||
[[Media:EMSegTutorial-AHM2008.ppt | Slides]] - [[Media:EMSegTutorial-AHM2008.zip | Data]] | [[Media:EMSegTutorial-AHM2008.ppt | Slides]] - [[Media:EMSegTutorial-AHM2008.zip | Data]] | ||
| + | |||
| + | * Export EMSEG_TUTORIAL_DIR environment variable ( e.g. <PATH TO TUTORIAL> = /tmp/EMSegmentTutorial-Jan2008/ ) | ||
| + | export EMSEG_TUTORIAL_DIR=<PATH_TO_TUTORIAL> | ||
===Quick Tour of Features and Use=== | ===Quick Tour of Features and Use=== | ||
| Line 47: | Line 50: | ||
The above EMSegment tutorial data set demonstrates the use of the command line interface. | The above EMSegment tutorial data set demonstrates the use of the command line interface. | ||
| − | If the command-line module is invoked from Slicer3 GUI, there will be a '''Parameters panel:''' [[Image:EMSegment-CL-Panel.png| | + | If the command-line module is invoked from Slicer3 GUI, there will be a '''Parameters panel:''' [[Image:EMSegment-CL-Panel.png|400px|thumb|EMSegment Simple Parameters Panel]] |
# MRML Scene: Select the mrml scene for the tutorial parameter set | # MRML Scene: Select the mrml scene for the tutorial parameter set | ||
# Result Labelmap: Select a new image filename for output labelmap | # Result Labelmap: Select a new image filename for output labelmap | ||
| Line 54: | Line 57: | ||
Here are some basic examples to run this module from command line directly | Here are some basic examples to run this module from command line directly | ||
| + | |||
| + | * Export the Slicer3_HOME environment variable ( e.g. <PATH_TO_SLICER> = /tmp/3DSlicer/Slicer3-build/ ) | ||
| + | export Slicer3_HOME=<PATH_TO_SLICER> | ||
* Run the executable to see the documentation | * Run the executable to see the documentation | ||
| − | $ | + | ${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine --help |
| − | *Run the segmentation algorithm with tutorial data | + | * Run the segmentation algorithm with tutorial data |
The script "Run-BrainTutorial-Defaults.sh" demonstrates how to run the EMSegmenter on the tutorial data. The resulting labelmap is written into the VolumeData_Output directory. Try adding the "--intermediateResultsDirectory myDirectoryName" flag to write out intermediate data. Intermediate data is saved in the "myDirectoryName" directory: you can load this scene to visualize the intermediate data (e.g. intensity normalized and registered images) and segmentation results. | The script "Run-BrainTutorial-Defaults.sh" demonstrates how to run the EMSegmenter on the tutorial data. The resulting labelmap is written into the VolumeData_Output directory. Try adding the "--intermediateResultsDirectory myDirectoryName" flag to write out intermediate data. Intermediate data is saved in the "myDirectoryName" directory: you can load this scene to visualize the intermediate data (e.g. intensity normalized and registered images) and segmentation results. | ||
| − | source | + | source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh |
RUN_NAME=BrainTutorial-Defaults | RUN_NAME=BrainTutorial-Defaults | ||
| − | $ | + | mkdir -p $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME |
| − | --verbose\ | + | ${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \ |
| − | --mrmlSceneFileName $EMSEG_TUTORIAL_DIR/EMSeg-Brain-MRT1T2.mrml\ | + | --verbose \ |
| − | --intermediateResultsDirectory $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME\ | + | --mrmlSceneFileName $EMSEG_TUTORIAL_DIR/EMSeg-Brain-MRT1T2.mrml \ |
| − | --resultVolumeFileName VolumeData_Output/Segmentation-$RUN_NAME.nhdr\ | + | --intermediateResultsDirectory $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME \ |
| + | --resultVolumeFileName $EMSEG_TUTORIAL_DIR/VolumeData_Output/Segmentation-$RUN_NAME.nhdr \ | ||
* Run the segmentation algorithm with new data | * Run the segmentation algorithm with new data | ||
| − | The other | + | The other script, "BrainTutorial-SetTarget1.sh" demonstrate how to run the EMSegmenter on new |
| − | data. | + | data. Because the tutorial EMSegment parameters are designed to accept one T1 and one T2 input image, each script supplies one new T1 and one T2 to be segmented (NB: order is important). |
| + | |||
| + | source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh | ||
| + | RUN_NAME=BrainTutorial-SetTarget | ||
| + | mkdir -p $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME | ||
| + | ${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \ | ||
| + | --verbose \ | ||
| + | --mrmlSceneFileName $EMSEG_TUTORIAL_DIR/EMSeg-Brain-MRT1T2.mrml \ | ||
| + | --targetVolumeFileNames \ | ||
| + | $EMSEG_TUTORIAL_DIR/VolumeData_Input/Target/t1.nhdr,$EMSEG_TUTORIAL_DIR/VolumeData_Input/Target/t2.nhdr \ | ||
| + | --resultVolumeFileName \ | ||
| + | VolumeData_Output/Segmentation-$RUN_NAME.nhdr \ | ||
| + | --intermediateResultsDirectory \ | ||
| + | $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME \ | ||
| + | |||
| + | * Run the MRI-Human-Brain task | ||
| − | + | source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh | |
| − | + | ${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \ | |
| − | + | --verbose \ | |
| − | --verbose\ | + | --mrmlSceneFileName \ |
| − | --mrmlSceneFileName $ | + | $Slicer3_HOME/share/Slicer3/Modules/EMSegment/Tasks/MRI-Human-Brain.mrml \ |
| − | -- | + | --targetVolumeFileNames \ |
| − | + | $Slicer3_HOME/../Slicer3/Modules/EMSegment/Testing/TestData/MiscVolumeData/MRIHumanBrain_T1_aligned.nrrd \ | |
| − | -- | + | --resultVolumeFileName \ |
| − | + | /tmp/Result_MRI-Human-Brain.nhdr \ | |
== Development == | == Development == | ||
| Line 101: | Line 123: | ||
* [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentBlackBoxSegmentationTest.cxx vtkEMSegmentBlackBoxSegmentationTest ] | * [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentBlackBoxSegmentationTest.cxx vtkEMSegmentBlackBoxSegmentationTest ] | ||
* [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentMRMLManagerTest.cxx vtkEMSegmentMRMLManagerTest ] | * [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentMRMLManagerTest.cxx vtkEMSegmentMRMLManagerTest ] | ||
| − | *[http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentPreprocessingManagerTest.cxx vtkEMSegmentPreprocessingManagerTest ] | + | * [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentPreprocessingManagerTest.cxx vtkEMSegmentPreprocessingManagerTest ] |
| − | *[http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentReadWriteMRMLTest.cxx vtkEMSegmentReadWriteMRMLTest ] | + | * [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentReadWriteMRMLTest.cxx vtkEMSegmentReadWriteMRMLTest ] |
| − | *[http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentTestUtilities.cxx vtkEMSegmentTestUtilities ] | + | * [http://viewvc.slicer.org/viewcvs.cgi/trunk/Modules/EMSegment/Testing/vtkEMSegmentTestUtilities.cxx vtkEMSegmentTestUtilities ] |
===Known bugs=== | ===Known bugs=== | ||
Latest revision as of 05:01, 30 November 2010
Home < Modules:EMSegment-Command-Line3.6Return to Slicer 3.6 Documentation
Return to EMSegmenter Overview Page
Module Name
EM Segment Command-Line Executable
General Information
Module Type & Category
Type: CLI
Category: Segmentation
Authors, Collaborators & Contact
- Sebastien Barre: Kitware, Inc.
- Brad Davis: Kitware, Inc.
- Kilian Pohl: IBM
- Polina Golland: MIT
- Yumin Yuan: Kitware, Inc.
- Contact: Kilian Pohl, pohl@csail.mit.edu
Module Description
This module is used to simplify the process of segmenting large collections of images by providing a command line interface to the EMSegment algorithm for script and batch processing.
Usage
The primary function of the EMSegmenter module is to step the user through the process of calibrating, via algorithm parameters, the segmentation algorithm to a particular set of input data. However, once a successful collection of parameters is established, the user will commonly want to bypass this detailed calibration process when segmenting new images by using those parameters collected from the calibration process in EMSegmenter module. The EMSegment Command-line module which wraps the EMSegmentCommandLine executable provides this batch processing capability. The EMSegment Command-line module is automatic generated from the EMSegmentCommandLine executable using the --xml option.
Use Cases, Examples, Tutorials
This module is especially appropriate for segmenting large quantity of data by scripted/batch processing. It requires predefined parameter set, and new target/atlas images can be specified on command line.
- A full Tutorial for all three EMSegment modules
These slides and data describe the Slicer3 implementation of all three EMSegment modules and demonstrate their use. A recent (later than 7Jan08) version of Slicer3 is required to run the test data.
- Export EMSEG_TUTORIAL_DIR environment variable ( e.g. <PATH TO TUTORIAL> = /tmp/EMSegmentTutorial-Jan2008/ )
export EMSEG_TUTORIAL_DIR=<PATH_TO_TUTORIAL>
Quick Tour of Features and Use
The above EMSegment tutorial data set demonstrates the use of the command line interface.
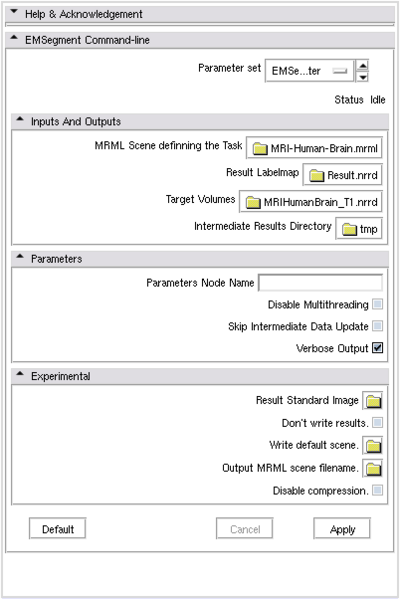
If the command-line module is invoked from Slicer3 GUI, there will be a Parameters panel:
- MRML Scene: Select the mrml scene for the tutorial parameter set
- Result Labelmap: Select a new image filename for output labelmap
- Target Volumes: Select input target volumes (Multiple files can be selected at one time)
- Click `Apply’
Here are some basic examples to run this module from command line directly
- Export the Slicer3_HOME environment variable ( e.g. <PATH_TO_SLICER> = /tmp/3DSlicer/Slicer3-build/ )
export Slicer3_HOME=<PATH_TO_SLICER>
- Run the executable to see the documentation
${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine --help
- Run the segmentation algorithm with tutorial data
The script "Run-BrainTutorial-Defaults.sh" demonstrates how to run the EMSegmenter on the tutorial data. The resulting labelmap is written into the VolumeData_Output directory. Try adding the "--intermediateResultsDirectory myDirectoryName" flag to write out intermediate data. Intermediate data is saved in the "myDirectoryName" directory: you can load this scene to visualize the intermediate data (e.g. intensity normalized and registered images) and segmentation results.
source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh
RUN_NAME=BrainTutorial-Defaults
mkdir -p $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME
${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \
--verbose \
--mrmlSceneFileName $EMSEG_TUTORIAL_DIR/EMSeg-Brain-MRT1T2.mrml \
--intermediateResultsDirectory $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME \
--resultVolumeFileName $EMSEG_TUTORIAL_DIR/VolumeData_Output/Segmentation-$RUN_NAME.nhdr \
- Run the segmentation algorithm with new data
The other script, "BrainTutorial-SetTarget1.sh" demonstrate how to run the EMSegmenter on new data. Because the tutorial EMSegment parameters are designed to accept one T1 and one T2 input image, each script supplies one new T1 and one T2 to be segmented (NB: order is important).
source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh
RUN_NAME=BrainTutorial-SetTarget
mkdir -p $EMSEG_TUTORIAL_DIR/Working/$RUN_NAME
${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \
--verbose \
--mrmlSceneFileName $EMSEG_TUTORIAL_DIR/EMSeg-Brain-MRT1T2.mrml \
--targetVolumeFileNames \
$EMSEG_TUTORIAL_DIR/VolumeData_Input/Target/t1.nhdr,$EMSEG_TUTORIAL_DIR/VolumeData_Input/Target/t2.nhdr \
--resultVolumeFileName \
VolumeData_Output/Segmentation-$RUN_NAME.nhdr \
--intermediateResultsDirectory \
$EMSEG_TUTORIAL_DIR/Working/$RUN_NAME \
- Run the MRI-Human-Brain task
source ${Slicer3_HOME}/bin/Slicer3SetupPaths.sh
${Slicer3_HOME}/Slicer3 --launch $Slicer3_HOME/lib/Slicer3/Plugins/EMSegmentCommandLine \
--verbose \
--mrmlSceneFileName \
$Slicer3_HOME/share/Slicer3/Modules/EMSegment/Tasks/MRI-Human-Brain.mrml \
--targetVolumeFileNames \
$Slicer3_HOME/../Slicer3/Modules/EMSegment/Testing/TestData/MiscVolumeData/MRIHumanBrain_T1_aligned.nrrd \
--resultVolumeFileName \
/tmp/Result_MRI-Human-Brain.nhdr \
Development
The EMSegment Command-line module is automatic generated from the EMSegmentCommandLine executable using the --xml option. The EMSegmentCommandLine provides a command line interface to the EMSegment algorithm for script/batch processing of large collection of data with predefined parameters set.
Notes from the Developer(s)
N/A
Dependencies
Slicer3 base modules
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- vtkEMSegmentBlackBoxSegmentationTest
- vtkEMSegmentMRMLManagerTest
- vtkEMSegmentPreprocessingManagerTest
- vtkEMSegmentReadWriteMRMLTest
- vtkEMSegmentTestUtilities
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
The EMSegmenter can be adapted to many segmentation problems. However, there is no "default" set of parameters that will work for all segmentation problems.
- Atlas-to-target registration and intensity normalization are very important; it will be most effective to apply these steps using algorithms that are customized to your data. Defaults are provided but they may perform poorly for your data---this will lead to poor segmentation results.
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links for the module.
More Information
- For more information about the EM Segmenter project see the EMSegmenter Wiki page here
Acknowledgment
Funding for the EMSegmenter module was provided by NAMIC and NAC.
References
Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212. [bib]