Difference between revisions of "Modules:EMDTIClustering-Documentation-3.6"
m |
|||
| Line 1: | Line 1: | ||
[[Documentation-3.6|Return to Slicer 3.6 Documentation]] | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] | ||
| − | [[Announcements:Slicer3. | + | [[Announcements:Slicer3.6#Highlights|Gallery of New Features]] |
__NOTOC__ | __NOTOC__ | ||
Revision as of 14:45, 4 May 2010
Home < Modules:EMDTIClustering-Documentation-3.6Return to Slicer 3.6 Documentation
Module Name
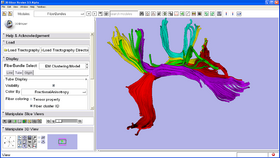
EM Fiber Clustering Module
General Information
Module Type & Category
Type: CLI
Category: Clustering
Authors, Collaborators & Contact
- Mahnaz Maddah
- James Miller
- Contact: Mahnaz Maddah, maddah@ge.com
Module Description
This module clusters a set of input trajectories into a number of bundles, generates arc length parameterization by establishing the point correspondences and reports diffusion parameters along the bundles as well as the membership probability of each trajectory in each cluster. The module requires specification of seed trajectories (or initial centerlines) as representatives of the desired bundles.
Usage
Examples, Use Cases & Tutorials
Tutorial and test data can be downloaded from here:
http://www.nitrc.org/projects/quantitativedti/
Quick Tour of Features and Use
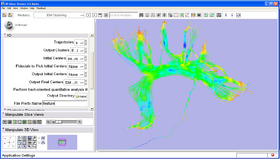
Here is a list of the panels in the module:
- IO panel:
Trajectories Input trajectories to be clustered.
Output Clusters Clustered trajecories labeled by their cluster ID.
Initial Centers [optional] Initial center(s). Note that intial centers need
to be provided by passing either a set of trajectories here or a fiducial list.
Fiducials to Pick Initial Centers [optional] A fiducial list to generate initial center(s). For each fiducial
in the list the closest trajectory in the input is selected as the initial center.
Output Initial Centers [Optional] Selected initial cluster centers.
Output Final Centers [Optional] Final cluster centers, colored by the mean FA value along the bundle
if "Perform Quantitative Analysis" is flaged .
Perform Quantitative Analysis Flag that needs to be marked if quantitative analysis is desired to be done.
Output Directory A directory needs to be specified if performing quantitative analysis.
File Prefix Name Prefix of the output files generated through tract-oriented analysis.
Description of generated CSV files by EM Clustering Module
- Clustering Parameters:
Compactness of Fiber Bundles Parameter between 1 and 5 that specifies the extent of similarity between the
trajectories of each cluster. Increase the value for more compact bundles.
- Advanced Parameters:
Space Resolution Space resolution for distance map calculation.
Iterations Maximum number of EM iterations.
Maximum Distance Maximum distance in mm specifies an upper threshold on the distance of points that
can contribute to new center formation.
Development
Notes from the Developer(s)
This module is based on the algorithms developped for quantitative analysis of white matter fiber tracts as a part of the author's PhD work: Mahnaz Maddah, Quantitative Analysis of Cerebral White Matter Anatomy from Diffusion MRI, Ph.D. Thesis CSAIL, MIT, September 2008.
Dependencies
NA
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- Test EMClusteringTest.cxx
Known bugs
Follow this link to the Slicer3 bug tracker.
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Source code link
More Information
Acknowledgment
This work has been supported by NAC.