Difference between revisions of "Modules:DiffusionMRIWelcome-Documentation-3.6"
| (43 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[Documentation-3.6|Return to Slicer 3.6 Documentation]] | ||
| + | |||
| + | __NOTOC__ | ||
| + | |||
= Diffusion MRI in 3D Slicer = | = Diffusion MRI in 3D Slicer = | ||
{| style="color:#000000; background-color:#aaaaaa;" cellpadding="10" cellspacing="0" border="0" | {| style="color:#000000; background-color:#aaaaaa;" cellpadding="10" cellspacing="0" border="0" | ||
| − | |An rich set of tools is available within 3D Slicer to perform Diffusion MRI image visualization and analysis. There are three categories of modules: [[ #DWI Filtering ]] for denoising the Diffusion Weighted (DW) images; [[ #Diffusion tensor utilities ]] for estimating diffusion tensors (DT) from DW images and calculating scalar invariants like the fractional anisotropy (FA) and resampling the DT images; and [[ #Tractography ]] to trace and analyse white matter fibers from DT images and assess connectivity between regions from DW images. | + | |An rich set of tools is available within 3D Slicer to perform Diffusion MRI image visualization and analysis. There are three categories of modules: [[ #DWI Filtering|DWI filtering ]] for denoising the Diffusion Weighted (DW) images; [[ #Diffusion Tensor Utilities|Diffusion tensor utilities ]] for estimating diffusion tensors (DT) from DW images and calculating scalar invariants like the fractional anisotropy (FA) and resampling the DT images; and [[ #Tractography|Tractography ]] to trace and analyse white matter fibers from DT images and assess connectivity between regions from DW images. |
| + | |||
| + | For a brief description of the common diffusion MRI processing pipeline please follow this [[ #Diffusion Processing Pipeline|link ]]. | ||
|} | |} | ||
| + | |||
| + | =Recommended Workflow in 3D Slicer= | ||
| + | ==1. Convert DWI data into nrrd format== | ||
| + | <big><big>'''As first step for handling your DICOM format DWI data, use the DicomToNrrd module to convert your DWI data from DICOM to NRRD format.'''</big></big> | ||
| + | * Then load the nrrd format data into Slicer using the File->Add Volume dialog. See [[Modules:DicomToNRRD-3.6|here]] for a '''list of supported DWI formats'''. | ||
| + | * See [[Slicer-3.6-Load-Overview|here]] for an overview over the process of loading data into Slicer. | ||
| + | |||
| + | ==2. Filter the DWI for noise reduction== | ||
| + | * Begin with the [[Modules:JointRicianLMMSEImageFilter-Documentation-3.6|Modules:Joint Rician LMMSE Image Filter]] | ||
| + | * Several other filters are available, but we have found this one to be easy to use and the defaults should work on human brains. | ||
| + | |||
| + | ==3. Estimate Tensors== | ||
| + | |||
| + | |||
| + | ==4. Perform Tractography== | ||
= Diffusion MRI Modules = | = Diffusion MRI Modules = | ||
| − | == DWI | + | == DWI Denoising== |
| + | [[Image:DenoisingIllustration.png|center|thumb|upright=4.|Result of the different DWI filtering algorithms. From left to right: Original images, | ||
| + | [[Modules:UnbiasedNonLocalMeans-Documentation-3.6|Unbiased Non Local Means filter for DWI]]; | ||
| + | [[Modules:RicianLMMSEImageFilter-Documentation-3.6|Rician LMMSE Image Filter]]; | ||
| + | [[Modules:JointRicianLMMSEImageFilter-Documentation-3.6|Joint Rician LMMSE Image Filter]]. Image from A. Tristán-Vega and S. Aja-Fernández. DWI filtering using joint information for DTI and HARDI. | ||
| + | Medical Image Analysis, 2009. | ||
| + | ]] | ||
| + | Three techniques are provided for denoising DW images: | ||
| + | *[[Modules:UnbiasedNonLocalMeans-Documentation-3.6|Unbiased Non Local Means filter for DWI]]: Is the one providing the most visually appealing results. However, it is very time consuming and may mix information from remote areas of the image. | ||
| + | *[[Modules:RicianLMMSEImageFilter-Documentation-3.6|Rician LMMSE Image Filter]]: Estimates Rician noise and uses this estimation and spatial coherence to perform the denoising. It processes each gradient direction individually. | ||
| + | *[[Modules:JointRicianLMMSEImageFilter-Documentation-3.6|Joint Rician LMMSE Image Filter]]: Estimates Rician noise and uses this estimation, spatial and orientational coherence to perform the denoising. It jointly processes several gradient directions. | ||
| + | |||
| + | == Diffusion Tensor Utilities== | ||
| + | {| | ||
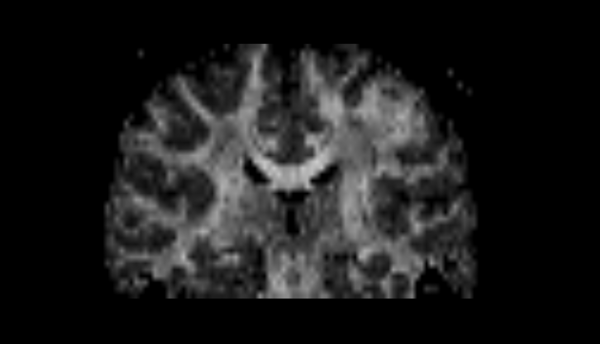
| + | |[[File:Tensor2.png|thumb|center|upright=2|Diffusion tensor image estimated from a DW image using the [[Modules:DiffusionTensorEstimation-Documentation-3.6|Diffusion Tensor Estimation]] module.]] | ||
| + | |[[File:Fa2.png|thumb|center|upright=2|Fractional anisotropy image calculated from a diffusion tensor image using the [[Modules:DiffusionTensorScalarMeasurements-Documentation-3.6 | Diffusion Tensor Scalar Measurements]] module.]] | ||
| + | |} | ||
| + | *[[Modules:DiffusionTensorEstimation-Documentation-3.6|Diffusion Tensor Estimation]]: Produces a diffusion tensor image from a DW image. | ||
| + | *[[Modules:DiffusionTensorScalarMeasurements-Documentation-3.6 | Diffusion Tensor Scalar Measurements]]: Calculates scalar invariants such as the fractional anisotropy (FA) or the linear measure (LM) from a diffusion tensor image. | ||
| + | *[[Modules:ResampleDTIVolume-Documentation-3.6|Resample DTI Volume]]: Increases or decreases the resolution of a diffusion tensor image. | ||
| + | |||
| + | == Tractography== | ||
| + | {| | ||
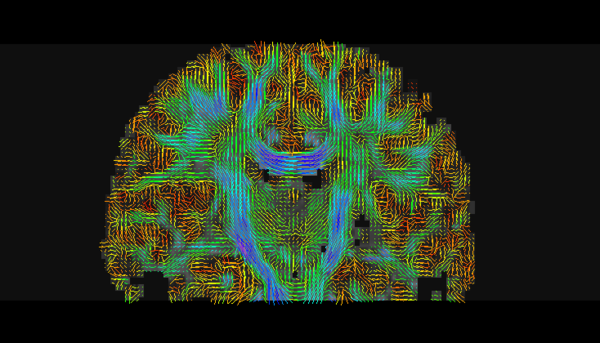
| + | |[[File:Roi tract.jpg|thumb|center|upright=2|Deterministic tractography result produced with the [[Modules:ROISeeding-Documentation-3.6 | Label Seeding]] or [[Modules:FiducialSeeding-Documentation-3.6|Fiducial Seeding]] modules.]]|| |[[File:General.png|thumb|center|upright=2|Stochastic tractography result produced with the [[Modules:StochasticTractography-Documentation-3.6|Stochastic Tractography]] module.]] | ||
| + | |} | ||
| + | *[[Modules:ROISeeding-Documentation-3.6 | Label Seeding]]: Deterministic tracing of the white matter fibers traversing a specified labeled region of the diffusion tensor image. | ||
| + | *[[Modules:FiducialSeeding-Documentation-3.6|Fiducial Seeding]]: Deterministic tracing of the white matter fibers traversing each [[Modules:Fiducials-Documentation-3.6|fiducial]] from a [[Modules:Fiducials-Documentation-3.6|fiducial list]]. | ||
| + | *[[Modules:DTIDisplay-Documentation-3.6|FiberBundles]]: Tuning of the visualization options for the deterministic tractography results produced with the [[Modules:ROISeeding-Documentation-3.6 | Label Seeding]] or [[Modules:FiducialSeeding-Documentation-3.6|Fiducial Seeding]] modules. | ||
| + | *[[Modules:StochasticTractography-Documentation-3.6|Stochastic Tractography]]: Probabilistically traces the white matter fibers connecting two regions. The output of this module is an image quantifying the probability that white matter fiber connecting two selected regions traverses each point in space. | ||
| + | *[[Modules:ROISelect-Documentation-3.6|ROI Select]]: Filters tracts produced by determinictractography passing through a region of interest expressed as a labeled region on the diffusion tensor image. | ||
| + | |||
| + | ==Diffusion Processing Pipeline== | ||
| + | ===First: Convert the Diffusion Weighted Images (DWI) from DICOM to NRRD=== | ||
| + | In order to use 3D Slicer to process the Diffusion Weighted Images obtained from a Diffusion Tensor imaging protocol, we must convert them to the NRRD format as follows: | ||
| + | [[Image:DWIDicomToNrrdGUI.png|thumb|280px|left|[[Modules:DicomToNRRD-3.6|'''Dicom To Nrrd''']] module]] | ||
| + | # Select the [[Modules:DicomToNRRD-3.6|'''Dicom To Nrrd''']] module from the '''Converters''' module category. | ||
| + | # Click on '''Input Dicom Data''' and select the directory where your the DICOM files corresponding to your DWI images are stored. | ||
| + | # Click on '''Output Directory''' and select the directory where you want to store the resulting NRRD file. | ||
| + | # Fill in '''Output Filename''' the name of the file, for instance: ''dwi.nrrd''. You must include the extension. | ||
| + | # Press the Apply button. | ||
| + | |||
| + | ===Second: Estimate the Diffusion Tensor Image (DT) from your DWI=== | ||
| + | We are now in position to estimate the Diffusion Tensor image from the DW image we have just calculated. However, in order to improve the quality of our image it is recommended to perform a denosing procedure beforehand. | ||
| + | ====Denoise the DW images==== | ||
| + | For this step, we recommend the [[Modules:JointRicianLMMSEImageFilter-Documentation-3.6|'''Joint Rician LMMSE Image Filter''']] due to its compromise of speed and soundness. This step is carried on by: | ||
| + | #Selecting the [[Modules:JointRicianLMMSEImageFilter-Documentation-3.6| '''Joint Rician LMMSE Image Filter''']] module which is under the '''Diffusion'' module category within the '''Denoising''' sub-category. | ||
| + | #Setting in the '''Input Volume''' field of the module the image obtained from the [[#First: Convert the Diffusion Weighted Images (DWI) from DICOM to NRRD | previous conversion step]]. For instance ''dwi.nhdr''. | ||
| + | #In the '''Output Volume''' field, selecting the option '''Create a New Diffusion Weighted Volume'''. This volume will have the automatically assigned name ''Joint Rician LMMSE Image Filter Volume''. | ||
| + | #Pressing the '''Apply''' button. | ||
| + | ====Estimate the DT from the DW image==== | ||
| + | Once we have the DW image we want, we use the [[Modules:DiffusionTensorEstimation-Documentation-3.6|'''Diffusion tensor Estimation''']] module located in the '''Diffusion''' module category to estimate the DT image from a DW: | ||
| + | |||
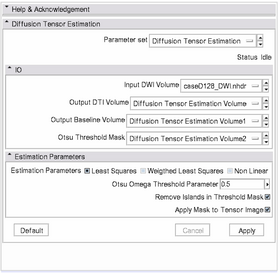
| + | [[Image:DiffusionTensorEstimationGUI.png|thumb|left|280px| [[Modules:DiffusionTensorEstimation-Documentation-3.6|'''Diffusion tensor Estimation''' module]] ]] | ||
| + | #Set the '''Input DWI Volume''' to the desired DW image. | ||
| + | #Set the fields | ||
| + | #;'''Output DTI Volume''' | ||
| + | #;'''Output Baseline Volume''' | ||
| + | #;'''Otsu Threshold Volume''' | ||
| + | #:to their corresponding ''Create new...'' option. You can then use the '''rename''' option if you do not like the default file names. | ||
| + | #Press the '''Apply''' button. | ||
| + | |||
| + | ===Third: Perform a tractography study=== | ||
Latest revision as of 02:55, 27 April 2011
Home < Modules:DiffusionMRIWelcome-Documentation-3.6Return to Slicer 3.6 Documentation
Diffusion MRI in 3D Slicer
| An rich set of tools is available within 3D Slicer to perform Diffusion MRI image visualization and analysis. There are three categories of modules: DWI filtering for denoising the Diffusion Weighted (DW) images; Diffusion tensor utilities for estimating diffusion tensors (DT) from DW images and calculating scalar invariants like the fractional anisotropy (FA) and resampling the DT images; and Tractography to trace and analyse white matter fibers from DT images and assess connectivity between regions from DW images.
For a brief description of the common diffusion MRI processing pipeline please follow this link . |
Recommended Workflow in 3D Slicer
1. Convert DWI data into nrrd format
As first step for handling your DICOM format DWI data, use the DicomToNrrd module to convert your DWI data from DICOM to NRRD format.
- Then load the nrrd format data into Slicer using the File->Add Volume dialog. See here for a list of supported DWI formats.
- See here for an overview over the process of loading data into Slicer.
2. Filter the DWI for noise reduction
- Begin with the Modules:Joint Rician LMMSE Image Filter
- Several other filters are available, but we have found this one to be easy to use and the defaults should work on human brains.
3. Estimate Tensors
4. Perform Tractography
Diffusion MRI Modules
DWI Denoising
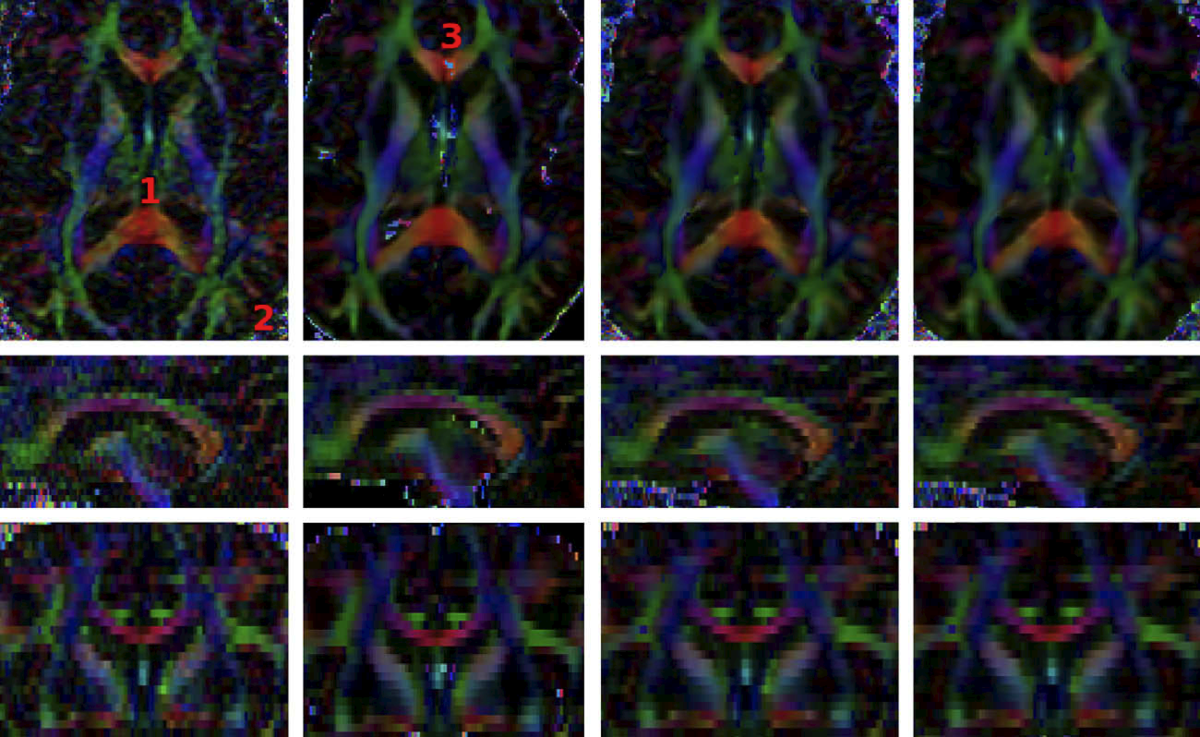
Three techniques are provided for denoising DW images:
- Unbiased Non Local Means filter for DWI: Is the one providing the most visually appealing results. However, it is very time consuming and may mix information from remote areas of the image.
- Rician LMMSE Image Filter: Estimates Rician noise and uses this estimation and spatial coherence to perform the denoising. It processes each gradient direction individually.
- Joint Rician LMMSE Image Filter: Estimates Rician noise and uses this estimation, spatial and orientational coherence to perform the denoising. It jointly processes several gradient directions.
Diffusion Tensor Utilities
 Diffusion tensor image estimated from a DW image using the Diffusion Tensor Estimation module. |
 Fractional anisotropy image calculated from a diffusion tensor image using the Diffusion Tensor Scalar Measurements module. |
- Diffusion Tensor Estimation: Produces a diffusion tensor image from a DW image.
- Diffusion Tensor Scalar Measurements: Calculates scalar invariants such as the fractional anisotropy (FA) or the linear measure (LM) from a diffusion tensor image.
- Resample DTI Volume: Increases or decreases the resolution of a diffusion tensor image.
Tractography
 Stochastic tractography result produced with the Stochastic Tractography module. |
- Label Seeding: Deterministic tracing of the white matter fibers traversing a specified labeled region of the diffusion tensor image.
- Fiducial Seeding: Deterministic tracing of the white matter fibers traversing each fiducial from a fiducial list.
- FiberBundles: Tuning of the visualization options for the deterministic tractography results produced with the Label Seeding or Fiducial Seeding modules.
- Stochastic Tractography: Probabilistically traces the white matter fibers connecting two regions. The output of this module is an image quantifying the probability that white matter fiber connecting two selected regions traverses each point in space.
- ROI Select: Filters tracts produced by determinictractography passing through a region of interest expressed as a labeled region on the diffusion tensor image.
Diffusion Processing Pipeline
First: Convert the Diffusion Weighted Images (DWI) from DICOM to NRRD
In order to use 3D Slicer to process the Diffusion Weighted Images obtained from a Diffusion Tensor imaging protocol, we must convert them to the NRRD format as follows:
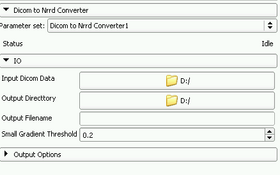
- Select the Dicom To Nrrd module from the Converters module category.
- Click on Input Dicom Data and select the directory where your the DICOM files corresponding to your DWI images are stored.
- Click on Output Directory and select the directory where you want to store the resulting NRRD file.
- Fill in Output Filename the name of the file, for instance: dwi.nrrd. You must include the extension.
- Press the Apply button.
Second: Estimate the Diffusion Tensor Image (DT) from your DWI
We are now in position to estimate the Diffusion Tensor image from the DW image we have just calculated. However, in order to improve the quality of our image it is recommended to perform a denosing procedure beforehand.
Denoise the DW images
For this step, we recommend the Joint Rician LMMSE Image Filter due to its compromise of speed and soundness. This step is carried on by:
- Selecting the Joint Rician LMMSE Image Filter module which is under the Diffusion module category within the Denoising' sub-category.
- Setting in the Input Volume field of the module the image obtained from the previous conversion step. For instance dwi.nhdr.
- In the Output Volume field, selecting the option Create a New Diffusion Weighted Volume. This volume will have the automatically assigned name Joint Rician LMMSE Image Filter Volume.
- Pressing the Apply button.
Estimate the DT from the DW image
Once we have the DW image we want, we use the Diffusion tensor Estimation module located in the Diffusion module category to estimate the DT image from a DW:
- Set the Input DWI Volume to the desired DW image.
- Set the fields
- Output DTI Volume
- Output Baseline Volume
- Otsu Threshold Volume
- to their corresponding Create new... option. You can then use the rename option if you do not like the default file names.
- Press the Apply button.