Difference between revisions of "Modules:DicomToNRRD-3.6"
(→Usage) |
|||
| Line 51: | Line 51: | ||
===Usability Issues=== | ===Usability Issues=== | ||
| − | + | Because different vendors used different private tags to store information related to diffusion weighting and documentation on how to interoperate this information is scarce, the current version support diffusion weighted images from GE, Siemens, and Philips scanners. For Siemens, both mosaic and split formats are supported. For Philips, both multi-slice and signal-slice formats are supported. | |
| + | |||
| + | The current version also support non-diffusion weighted images from any vendors (that GDCM supports), in which case, the module acts as a dicom series to nrrd volume converter. | ||
===Source Code and Documentation=== | ===Source Code and Documentation=== | ||
Revision as of 15:26, 26 April 2010
Home < Modules:DicomToNRRD-3.6Return to Slicer 3.6 Documentation
Module Name
DWI Dicom To NRRD
General Information
Module Type & Category
Type: Command line module
Category: Converters
Authors, Collaborators & Contact
- Author: Xiaodong Tao (with contribution from Vince Magnotta and Hans Johnson)
- Contact: taox @ research.ge.com
Module Description
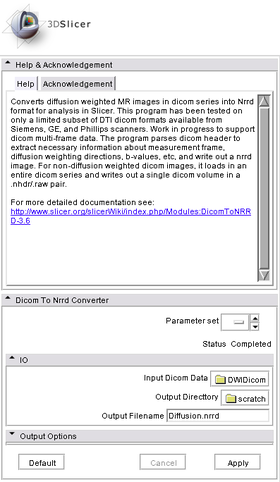
This module converts diffusion weighted MR images in dicom series into Nrrd format for analysis in Slicer. This program has been tested on only a limited subset of DTI dicom formats available from Siemens, GE, and Phillips scanners. Work in progress to support dicom multi-frame data. The program parses dicom header to extract necessary information about measurement frame, diffusion weighting directions, b-values, etc, and write out a nrrd image.
Usage
Examples, Use cases & Tutorials
Quick Tour of Features and Use
When this module is run from Slicer UI, simply select the "DICOM Directory" and "Output Image Volume" and press "Apply" button.
The module can also be run from command line to enable batch process. An example command line look like:
d:\Builds\Slicer3\lib\Slicer3\Plugins\Release\DicomToNrrdConverter.exe --inputDicomDirectory MyDicomDir --outputVolume MyNrrdImage.nhdr
Development
Dependencies
This module depends only on the core "Volumes" module of slice for data IO.
Known Bugs
Usability Issues
Because different vendors used different private tags to store information related to diffusion weighting and documentation on how to interoperate this information is scarce, the current version support diffusion weighted images from GE, Siemens, and Philips scanners. For Siemens, both mosaic and split formats are supported. For Philips, both multi-slice and signal-slice formats are supported.
The current version also support non-diffusion weighted images from any vendors (that GDCM supports), in which case, the module acts as a dicom series to nrrd volume converter.
Source Code and Documentation
Source Code: DicomToNrrdConverter.cxx
XML Description: DicomToNrrdConverter.xml
./DicomToNrrdConverter --help
USAGE:
d:\Builds\Slicer3\lib\Slicer3\Plugins\Release\DicomToNrrdConverter.exe
[--processinformationaddress
<std::string>] [--xml] [--echo]
[--useIdentityMeaseurementFrame]
[--writeProtocolGradientsFile]
[--useLPS] [--outputVolume
<std::string>]
[--inputDicomDirectory
<std::string>] [--] [--version]
[-h]
Where:
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--useIdentityMeaseurementFrame (advanced option, use default)
Adjust all the gradients so that the measurement frame is an identity
matrix. (default: 0)
--writeProtocolGradientsFile (advanced option, for debug)
Write the protocol gradients to a file suffixed by '.txt' as they were
specified in the procol by multiplying each diffusion gradient
direction by the measurement frame. This file is for debugging
purposes only, the format is not fixed, and will likely change as
debugging of new dicom formats is necessary. (default: 0)
--useLPS (advanced option, use default)
For GE data only. Specifies that the output should be in LPS space
like the original DICOM data. The default is to use Slicer3
consistent RAS space. (default: 0)
--outputVolume <std::string>
Output Image in NRRD format (.nhdr)
--inputDicomDirectory <std::string>
Directory holding Dicom series
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
Description: Converts diffusion weighted MR images in dicom series into Nrrd
format for analysis in Slicer. This program has been tested on only
a limited subset of DTI dicom formats available from Siemens, GE,
and Phillips scanners. Work in progress to support dicom multi-frame
data. The program parses dicom header to extract necessary
information about measurement frame, diffusion weighting directions,
b-values, etc, and write out a nrrd image.
Author(s): Xiaodong Tao
Acknowledgements: This work is part of the National Alliance for Medical
Image Computing (NAMIC), funded by the National Institutes of Health
through the NIH Roadmap for Medical Research, Grant U54 EB005149.
Additional support for DTI data produced on Philips scanners was
contributed by Vincent Magnotta and Hans Johnson at the University of
Iowa.
More Information
Acknowledgement
Acknowledgements: This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Additional support for DTI data produced on Philips scanners was contributed by Vincent Magnotta and Hans Johnson at the University of Iowa.