Difference between revisions of "Modules:BRAINSFit"
Hjmjohnson (talk | contribs) |
Hjmjohnson (talk | contribs) |
||
| Line 17: | Line 17: | ||
Category: Registration | Category: Registration | ||
| + | |||
| + | |||
===Authors, Collaborators & Contact=== | ===Authors, Collaborators & Contact=== | ||
| − | + | ||
| − | + | Author: Hans J. Johnson, hans-johnson -at- uiowa.edu, http://wwww.psychiatry.uiowa.edu | |
| − | + | ||
| + | Contributors: Hans Johnson(1,3,4); Kent Williams(1); Gregory Harris(1), Vincent Magnotta(1,2,3); Andriy Fedorov(5), fedorov -at- bwh.harvard.edu (Slicer integration); (1=University of Iowa Department of Psychiatry, 2=University of Iowa Department of Radiology, 3=University of Iowa Department of Biomedical Engineering, 4=University of Iowa Department of Electrical and Computer Engineering, 5=Surgical Planning Lab, Harvard) | ||
| + | |||
| + | |||
===Module Description=== | ===Module Description=== | ||
| − | BRAINSFit | + | |
| + | {| style="color:green" border="1" | ||
| + | |Program title || BRAINSFit | ||
| + | |||
| + | |- | ||
| + | |Program version || 3.0.0 | ||
| + | |||
| + | |- | ||
| + | |Program documentation-url || http://wiki.slicer.org/slicerWiki/index.php/Modules:BRAINSFit | ||
| + | |||
| + | |- | ||
| + | |} | ||
Insight/Examples/Registration/ImageRegistration8.cxx | Insight/Examples/Registration/ImageRegistration8.cxx | ||
Revision as of 20:56, 4 June 2010
Home < Modules:BRAINSFitReturn to Slicer 3.6 Documentation
Module Name
BRAINSFit
General Information
Module Type & Category
Type: CLI
Category: Registration
Authors, Collaborators & Contact
Author: Hans J. Johnson, hans-johnson -at- uiowa.edu, http://wwww.psychiatry.uiowa.edu
Contributors: Hans Johnson(1,3,4); Kent Williams(1); Gregory Harris(1), Vincent Magnotta(1,2,3); Andriy Fedorov(5), fedorov -at- bwh.harvard.edu (Slicer integration); (1=University of Iowa Department of Psychiatry, 2=University of Iowa Department of Radiology, 3=University of Iowa Department of Biomedical Engineering, 4=University of Iowa Department of Electrical and Computer Engineering, 5=Surgical Planning Lab, Harvard)
Module Description
| Program title | BRAINSFit |
| Program version | 3.0.0 |
| Program documentation-url | http://wiki.slicer.org/slicerWiki/index.php/Modules:BRAINSFit |
Insight/Examples/Registration/ImageRegistration8.cxx This program is the most functional example of multi-modal 3D rigid image registration provided with ITK. ImageRegistration8 is in the Examples directory, and also sec. 8.5.3 in the ITK manual. We have modified and extended this example in several ways:
- defined a new ITK Transform class, based on itkScaleSkewVersor3DTransform which has 3 dimensions of scale but no skew aspect.
- implemented a set of functions to convert between Versor Transforms and the general itk::AffineTransform and deferred converting from specific to more general representations to preserve transform information specificity as long as possible. Our Rigid transform is the narrowest, a Versor rotation plus separate translation.
- Added a template class itkMultiModal3DMutualRegistrationHelper which is templated over the type of ITK transform generated, and the optimizer used.
- Added image masks as an optional input to the Registration algorithm, limiting the volume considered during registration to voxels within the brain.
- Added image mask generation as an optional input to the Registration algorithm when meaningful masks such as for whole brain are not available, allowing the fit to at least be focused on whole head tissue.
- Added the ability to use one transform result, such as the Rigid transform, to initialize a more adaptive transform
- Defined the command line parameters using tools from the Slicer [ 3] program, in order to conform to the Slicer3 Execution model.
Added the ability to write output images in any ITK-supported scalar image format.
- Extensive testing as part of the BRAINS2 application suite, determined reasonable defaults for registration algorithm parameters. http://testing.psychiatry.uiowa.edu/CDash/
Usage
The BRAINSFit distribution contains a directory named TestData, which contains two example images. The first, test.nii.gz is a NIfTI format image volume, which is used the input for the CTest-managed regression test program. The program makexfrmedImage.cxx, included in the BRAINSFit distribution was used to generate test2.nii.gz, by scaling, rotating and translating test.nii.gz. You can see representative Sagittal slices of test.nii.gz (in this case, the fixed image, test2.nii.gz (the moving image), and the two images ’checkerboarded’ together to the right. To register test2.nii.gz to test.nii.gz, you can use the following command:
BRAINSFit --fixedVolume test.nii.gz \ --movingVolume test2.nii.gz \ --outputVolume registered.nii.gz \ --transformType Affine
A representative slice of the registered results image registered.nii.gz is to the right. You can see from the Checkerboard of the Fixed and Registered images that the fit is quite good with Affine transform-based registration. The blurring of the registered images is a consequence of the initial scaling used in generating the moving image from the fixed image, compounded by the interpolation necessitated by the transform operation.
You can see the differences in results if you re-run BRAINSFit using Rigid, ScaleVersor3D, or ScaleSkewVersor3D as the ----transformType parameter. In this case, the authors judged Affine the best method for registering these particular two images; in the BRAINS2 automated processing pipeline, Rigid usually works well for registering research scans.
Quick Tour of Features and Use
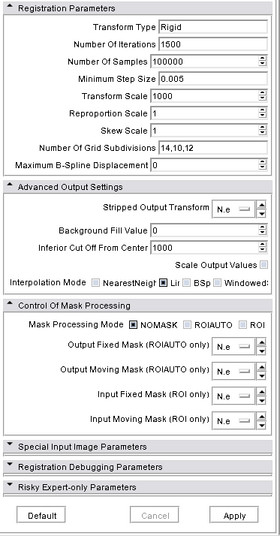
A list panels in the interface, their features, what they mean, and how to use them. For instance:
|
- Infererior Cut Off From Center: the cut-off below the image centers, in millimeters, (default: 1000)
Development
Notes from the Developer(s)
Dependencies
BRAINSFit depends on Slicer3 (for the SlicerExecutionModel support) and ITK.
Tests
TODO: Link to BRAINS3 and/or Slicer3 dashboard tests.
Known bugs
Links to known bugs in the Slicer3 bug tracker
Usability issues
Follow this link to the Slicer3 bug tracker. Please select the usability issue category when browsing or contributing.
Source code & documentation
Links to the module's source code:
Source code:
Doxygen documentation:
More Information
Acknowledgment
Include funding and other support here.
References
Publications related to this module go here. Links to pdfs would be useful.