Difference between revisions of "Modules:AtlasCreator:CongealingCLI"
m (Text replacement - "https?:\/\/(?:www|wiki)\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) " to "https://www.slicer.org/wiki/$1 ") |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 14: | Line 14: | ||
Author: Daniel Haehn and Kilian Pohl, University of Pennsylvania | Author: Daniel Haehn and Kilian Pohl, University of Pennsylvania | ||
| − | + | Collaborators: J. De Bonet, L. Zöllei and W.M. Wells III | |
| − | + | ||
| + | Acknowledgment: The research was funded by an ARRA supplement to NIH NCRR (P41 RR13218). | ||
| + | |||
== Graphical User Interface in 3D Slicer == | == Graphical User Interface in 3D Slicer == | ||
{| | {| | ||
| Line 35: | Line 37: | ||
|- | |- | ||
| − | |Program documentation-url || | + | |Program documentation-url || https://www.slicer.org/wiki/Modules:AtlasCreator |- |
| − | |||
| − | |- | ||
|} | |} | ||
| Line 103: | Line 103: | ||
./CongealingCLI [--returnparameterfile <std::string>] | ./CongealingCLI [--returnparameterfile <std::string>] | ||
[--processinformationaddress <std::string>] [--xml] | [--processinformationaddress <std::string>] [--xml] | ||
| − | [--echo] [--launch <std::string>] [-- | + | [--echo] [--launch <std::string>] [--outputPath |
| − | <std::string>] [--congeal_optimize_bestpoints <int>] | + | <std::string>] [--test <std::string>] |
| + | [--congeal_optimize_bestpoints <int>] | ||
[--congeal_schedule__n__optimize_samples | [--congeal_schedule__n__optimize_samples | ||
<std::vector<int>>] | <std::vector<int>>] | ||
| Line 174: | Line 175: | ||
The path to the congeal executable. Congeal will only be executed, if | The path to the congeal executable. Congeal will only be executed, if | ||
this is set. | this is set. | ||
| + | |||
| + | --outputPath <std::string> | ||
| + | The output path for the congeal configuration file. The file will only | ||
| + | be written, if this is set. (default: /tmp/congeal.config) | ||
--test <std::string> | --test <std::string> | ||
| Line 361: | Line 366: | ||
<pre> | <pre> | ||
$ ./CongealingCLI | $ ./CongealingCLI | ||
| − | Configuration file written to | + | Configuration file written to /tmp/congealing.config |
</pre> | </pre> | ||
| Line 380: | Line 385: | ||
--congeal_schedule__n__optimize_warp__3__ false,false,false,false,true --congeal_schedule__n__optimize_iterations 30,30,30,30,30 | --congeal_schedule__n__optimize_warp__3__ false,false,false,false,true --congeal_schedule__n__optimize_iterations 30,30,30,30,30 | ||
--congeal_schedule__n__optimize_samples 50000,50000,500000,500000,500000 --congeal_optimize_bestpoints 1000 --test congeal | --congeal_schedule__n__optimize_samples 50000,50000,500000,500000,500000 --congeal_optimize_bestpoints 1000 --test congeal | ||
| − | Configuration file written to | + | Configuration file written to /tmp/congealing.config |
</pre> | </pre> | ||
Latest revision as of 13:19, 27 November 2019
Home < Modules:AtlasCreator:CongealingCLIContents
Congealing Commandline Wrapper
CongealingCLI is a wrapper to access the Congealing Un-biased Groupwise Registration tool. It is now possible to generate a configuration file for Congealing by using a GUI or command line arguments. In fact, by not specifying any arguments and just running the wrapper, a default configuration file for congeal is generated.
Module Type & Category
Type: CLI
Category: Registration
Authors, Collaborators & Contact
Author: Daniel Haehn and Kilian Pohl, University of Pennsylvania
Collaborators: J. De Bonet, L. Zöllei and W.M. Wells III
Acknowledgment: The research was funded by an ARRA supplement to NIH NCRR (P41 RR13218).
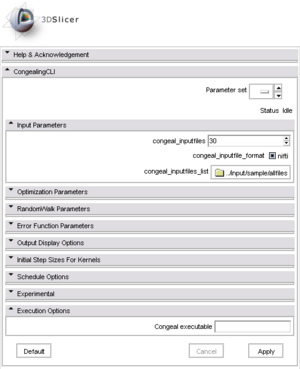
Graphical User Interface in 3D Slicer
Module Description
| Program title | CongealingCLI
|
| Program version | 0.1.1 |
| Program documentation-url | - |
- Input parameters The input parameters for Congealing.
- congeal_inputfiles [----congeal_inputfiles] : number of input files to use. Use '0' for all files when used in conjuctions with congeal_inputfiles.list
- congeal_inputfile_format [----congeal_inputfile_format] : format of input files. Currently only 'nifti' is supported
- congeal_inputfiles_list [----congeal_inputfiles_list] : a path to a file containing a list of input data files. The list file should contain one filename per line. Only congeal_inputfiles files will be used as input unless congeal_inputfiles is set to '0' in which case all the files in the list will be used
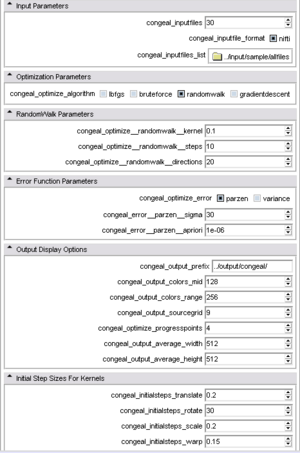
- Optimization parameters Options to configure the optimization.
- congeal_optimize_algorithm [----congeal_optimize_algorithm] : determines the optimization algorithm used
- RandomWalk parameters Options to configure RandomWalk.
- congeal_optimize__randomwalk__kernel [----congeal_optimize_randomwalk_kernel] : size of support to use for computing initial stepsize. This factor is multiplied by *.initialsteps to establish a maximum step radius for each dimensions
- congeal_optimize__randomwalk__steps [----congeal_optimize__randomwalk__steps] : maximum number of steps to take along any beam
- congeal_optimize__randomwalk__directions [----congeal_optimize__randomwalk__directions] : number of beams to try during each iteration
- Error function parameters Options to configure the error function
- congeal_optimize_error [----congeal_optimize_error] : selects the error metric to be used. parzen -- entropy estimate based on Parzen density estimator. variance -- variance of voxel stack.
- congeal_error__parzen__sigma [----congeal_error__parzen__sigma] : sigma of Gaussian used as kernel in Parzen density estimator. Measured in voxel intensity
- congeal_error__parzen__apriori [----congeal_error__parzen__apriori] : constant factor added to each Parzen estimate
- Output display options Options to configure the output display
- congeal_output_prefix [----congeal_output_prefix] : string prepended to the filenames of the outputfiles. This value can include an absolute or relative path, as well as a file prefix
- congeal_output_colors_mid [----congeal_output_colors_mid] : color equalization intercept. This value determines which data value will be mapped to mid gray
- congeal_output_colors_range [----congeal_output_colors_range] : color equalization slope. This value determines the relationship between changes in data value and changes in output image gray value
- congeal_output_sourcegrid [----congeal_output_sourcegrid] : determines how many of the transformed source values are shown in the *-inputs* images
- congeal_optimize_progresspoints [----congeal_optimize_progresspoints] : determines how many output file sets will be generated during each schedule
- congeal_output_average_width [----congeal_output_average_width] : determines the width of the congealing average visualization
- congeal_output_average_height [----congeal_output_average_height] : determines the height of the congealing average visualization
- Initial step sizes for kernels Option to configure the initial kernels
- congeal_initialsteps_translate [----congeal_initialsteps_translate] : relative scaling of translation parameters when computing kernels and step sizes. Scale: translation as fraction of image size
- congeal_initialsteps_rotate [----congeal_initialsteps_rotate] : relative scaling of rotation parameters when computing kernels and step sizes. Scale: rotation in degrees
- congeal_initialsteps_scale [----congeal_initialsteps_scale] : relative scaling of scaling parameters when computing kernels and step sizes. Scale: Scale as fraction of image size
- congeal_initialsteps_warp [----congeal_initialsteps_warp] : relative scaling of warp control point displacement when computing kernels and step sizes. Scale: Warp as fraction of control point's region
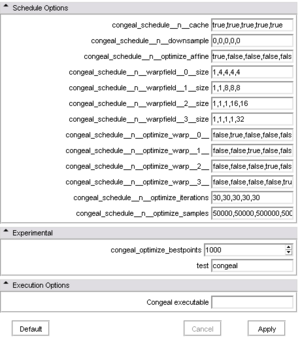
- Schedule options Options to configure the schedules
- congeal_schedule__n__cache [----congeal_schedule__n__cache] : determines whether or not the schedules results can be retrieved from the previous run. Separated by comma for each schedule run.
- congeal_schedule__n__downsample [----congeal_schedule__n__downsample] : determines how many times the input data should be downsampled (by factor of 2 in each dimension) prior to congealing. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_affine [----congeal_schedule__n__optimize_affine] : determines if affine parameters should be optimized or left fixed. Separated by comma for each schedule run.
- congeal_schedule__n__warpfield__0__size [----congeal_schedule__n__warpfield__0__size] : determines number of support points in each dimension of of B-Spline mesh for field 0. Separated by comma for each schedule run.
- congeal_schedule__n__warpfield__1__size [----congeal_schedule__n__warpfield__1__size] : determines number of support points in each dimension of of B-Spline mesh for field 1. Separated by comma for each schedule run.
- congeal_schedule__n__warpfield__2__size [----congeal_schedule__n__warpfield__2__size] : determines number of support points in each dimension of of B-Spline mesh for field 2. Separated by comma for each schedule run.
- congeal_schedule__n__warpfield__3__size [----congeal_schedule__n__warpfield__3__size] : determines number of support points in each dimension of of B-Spline mesh for field 3. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_warp__0__ [----congeal_schedule__n__optimize_warp__0__] : determines if the B-spline parameters for B-spline field 0 should be optimized or left fixed. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_warp__1__ [----congeal_schedule__n__optimize_warp__1__] : determines if the B-spline parameters for B-spline field 1 should be optimized or left fixed. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_warp__2__ [----congeal_schedule__n__optimize_warp__2__] : determines if the B-spline parameters for B-spline field 2 should be optimized or left fixed. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_warp__3__ [----congeal_schedule__n__optimize_warp__3__] : determines if the B-spline parameters for B-spline field 3 should be optimized or left fixed. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_iterations [----congeal_schedule__n__optimize_iterations] : number of optimzation iterations to be taken in the schedules. Separated by comma for each schedule run.
- congeal_schedule__n__optimize_samples [----congeal_schedule__n__optimize_samples] : number of samples to be compared in each transformed input volume. Separated by comma for each schedule run.
- Experimental Experimental options
- congeal_optimize_bestpoints [----congeal_optimize_bestpoints] :
- test [----test] : Currently unused. "Must be congeal."
- Write and Execution options Options to configure the execution
- Output path for configuration [----outputPath] : The output path for the congeal configuration file. The file will only be written, if this is set.
- Congeal executable [----launch] : The path to the congeal executable. Congeal will only be executed, if this is set.
Command Line Interface
The option --help prints the possible command line arguments:
$ ./CongealingCLI --help
USAGE:
./CongealingCLI [--returnparameterfile <std::string>]
[--processinformationaddress <std::string>] [--xml]
[--echo] [--launch <std::string>] [--outputPath
<std::string>] [--test <std::string>]
[--congeal_optimize_bestpoints <int>]
[--congeal_schedule__n__optimize_samples
<std::vector<int>>]
[--congeal_schedule__n__optimize_iterations
<std::vector<int>>]
[--congeal_schedule__n__optimize_warp__3__
<std::vector<std::string>>]
[--congeal_schedule__n__optimize_warp__2__
<std::vector<std::string>>]
[--congeal_schedule__n__optimize_warp__1__
<std::vector<std::string>>]
[--congeal_schedule__n__optimize_warp__0__
<std::vector<std::string>>]
[--congeal_schedule__n__warpfield__3__size
<std::vector<int>>]
[--congeal_schedule__n__warpfield__2__size
<std::vector<int>>]
[--congeal_schedule__n__warpfield__1__size
<std::vector<int>>]
[--congeal_schedule__n__warpfield__0__size
<std::vector<int>>]
[--congeal_schedule__n__optimize_affine
<std::vector<std::string>>]
[--congeal_schedule__n__downsample <std::vector<int>>]
[--congeal_schedule__n__cache
<std::vector<std::string>>]
[--congeal_initialsteps_warp <float>]
[--congeal_initialsteps_scale <float>]
[--congeal_initialsteps_rotate <float>]
[--congeal_initialsteps_translate <float>]
[--congeal_output_average_height <int>]
[--congeal_output_average_width <int>]
[--congeal_optimize_progresspoints <int>]
[--congeal_output_sourcegrid <int>]
[--congeal_output_colors_range <int>]
[--congeal_output_colors_mid <int>]
[--congeal_output_prefix <std::string>]
[--congeal_error__parzen__apriori <float>]
[--congeal_error__parzen__sigma <float>]
[--congeal_optimize_error <parzen|variance>]
[--congeal_optimize__randomwalk__directions <int>]
[--congeal_optimize__randomwalk__steps <int>]
[--congeal_optimize_randomwalk_kernel <float>]
[--congeal_optimize_algorithm <lbfgs|bruteforce
|randomwalk|gradientdescent>]
[--congeal_inputfiles_list <std::string>]
[--congeal_inputfile_format <nifti>]
[--congeal_inputfiles <int>] [--] [--version] [-h]
Where:
--returnparameterfile <std::string>
Filename in which to write simple return parameters (int, float,
int-vector, etc.) as opposed to bulk return parameters (image,
geometry, transform, measurement, table).
--processinformationaddress <std::string>
Address of a structure to store process information (progress, abort,
etc.). (default: 0)
--xml
Produce xml description of command line arguments (default: 0)
--echo
Echo the command line arguments (default: 0)
--launch <std::string>
The path to the congeal executable. Congeal will only be executed, if
this is set.
--outputPath <std::string>
The output path for the congeal configuration file. The file will only
be written, if this is set. (default: /tmp/congeal.config)
--test <std::string>
Currently unused. 'Must be congeal.' (default: congeal)
--congeal_optimize_bestpoints <int>
(default: 1000)
--congeal_schedule__n__optimize_samples <std::vector<int>>
number of samples to be compared in each transformed input volume.
Separated by comma for each schedule run. (default: 50000,50000,500000
,500000,500000)
--congeal_schedule__n__optimize_iterations <std::vector<int>>
number of optimzation iterations to be taken in the schedules.
Separated by comma for each schedule run. (default: 30,30,30,30,30)
--congeal_schedule__n__optimize_warp__3__ <std::vector<std::string>>
determines if the B-spline parameters for B-spline field 3 should be
optimized or left fixed. Separated by comma for each schedule run.
(default: false,false,false,false,true)
--congeal_schedule__n__optimize_warp__2__ <std::vector<std::string>>
determines if the B-spline parameters for B-spline field 2 should be
optimized or left fixed. Separated by comma for each schedule run.
(default: false,false,false,true,false)
--congeal_schedule__n__optimize_warp__1__ <std::vector<std::string>>
determines if the B-spline parameters for B-spline field 1 should be
optimized or left fixed. Separated by comma for each schedule run.
(default: false,false,true,false,false)
--congeal_schedule__n__optimize_warp__0__ <std::vector<std::string>>
determines if the B-spline parameters for B-spline field 0 should be
optimized or left fixed. Separated by comma for each schedule run.
(default: false,true,false,false,false)
--congeal_schedule__n__warpfield__3__size <std::vector<int>>
determines number of support points in each dimension of of B-Spline
mesh for field 3. Separated by comma for each schedule run. (default:
1,1,1,1,32)
--congeal_schedule__n__warpfield__2__size <std::vector<int>>
determines number of support points in each dimension of of B-Spline
mesh for field 2. Separated by comma for each schedule run. (default:
1,1,1,16,16)
--congeal_schedule__n__warpfield__1__size <std::vector<int>>
determines number of support points in each dimension of of B-Spline
mesh for field 1. Separated by comma for each schedule run. (default:
1,1,8,8,8)
--congeal_schedule__n__warpfield__0__size <std::vector<int>>
determines number of support points in each dimension of of B-Spline
mesh for field 0. Separated by comma for each schedule run. (default:
1,4,4,4,4)
--congeal_schedule__n__optimize_affine <std::vector<std::string>>
determines if affine parameters should be optimized or left fixed.
Separated by comma for each schedule run. (default: true,false,false
,false,false)
--congeal_schedule__n__downsample <std::vector<int>>
determines how many times the input data should be downsampled (by
factor of 2 in each dimension) prior to congealing. Separated by comma
for each schedule run. (default: 0,0,0,0,0)
--congeal_schedule__n__cache <std::vector<std::string>>
determines whether or not the schedules results can be retrieved from
the previous run. Separated by comma for each schedule run. (default:
true,true,true,true,true)
--congeal_initialsteps_warp <float>
relative scaling of warp control point displacement when computing
kernels and step sizes. Scale: Warp as fraction of control point's
region (default: 0.15)
--congeal_initialsteps_scale <float>
relative scaling of scaling parameters when computing kernels and step
sizes. Scale: Scale as fraction of image size (default: 0.2)
--congeal_initialsteps_rotate <float>
relative scaling of rotation parameters when computing kernels and
step sizes. Scale: rotation in degrees (default: 30)
--congeal_initialsteps_translate <float>
relative scaling of translation parameters when computing kernels and
step sizes. Scale: translation as fraction of image size (default:
0.2)
--congeal_output_average_height <int>
determines the height of the congealing average visualization
(default: 512)
--congeal_output_average_width <int>
determines the width of the congealing average visualization (default:
512)
--congeal_optimize_progresspoints <int>
determines how many output file sets will be generated during each
schedule (default: 4)
--congeal_output_sourcegrid <int>
determines how many of the transformed source values are shown in the
*-inputs* images (default: 9)
--congeal_output_colors_range <int>
color equalization slope. This value determines the relationship
between changes in data value and changes in output image gray value
(default: 256)
--congeal_output_colors_mid <int>
color equalization intercept. This value determines which data value
will be mapped to mid gray (default: 128)
--congeal_output_prefix <std::string>
string prepended to the filenames of the outputfiles. This value can
include an absolute or relative path, as well as a file prefix
(default: ../output/congeal/)
--congeal_error__parzen__apriori <float>
constant factor added to each Parzen estimate (default: 1e-06)
--congeal_error__parzen__sigma <float>
sigma of Gaussian used as kernel in Parzen density estimator. Measured
in voxel intensity (default: 30)
--congeal_optimize_error <parzen|variance>
selects the error metric to be used. parzen -- entropy estimate based
on Parzen density estimator. variance -- variance of voxel stack.
(default: parzen)
--congeal_optimize__randomwalk__directions <int>
number of beams to try during each iteration (default: 20)
--congeal_optimize__randomwalk__steps <int>
maximum number of steps to take along any beam (default: 10)
--congeal_optimize_randomwalk_kernel <float>
size of support to use for computing initial stepsize. This factor is
multiplied by *.initialsteps to establish a maximum step radius for
each dimensions (default: 0.1)
--congeal_optimize_algorithm <lbfgs|bruteforce|randomwalk
|gradientdescent>
determines the optimization algorithm used (default: randomwalk)
--congeal_inputfiles_list <std::string>
a path to a file containing a list of input data files. The list file
should contain one filename per line. Only congeal_inputfiles files
will be used as input unless congeal_inputfiles is set to '0' in which
case all the files in the list will be used (default:
../input/sample/allfiles)
--congeal_inputfile_format <nifti>
format of input files. Currently only 'nifti' is supported (default:
nifti)
--congeal_inputfiles <int>
number of input files to use. Use '0' for all files when used in
conjuctions with congeal_inputfiles.list (default: 30)
--, --ignore_rest
Ignores the rest of the labeled arguments following this flag.
--version
Displays version information and exits.
-h, --help
Displays usage information and exits.
Description: Generates a configuration file for the Congealing
Registration tool.
Author(s): Daniel Haehn and Kilian Pohl, University of
Pennsylvania
Acknowledgements: The research was funded by an ARRA supplement to NIH
NCRR (P41 RR13218).
Examples
The following commands are possible:
$ ./CongealingCLI Configuration file written to /tmp/congealing.config
or
$ ./CongealingCLI --congeal_inputfiles 30 --congeal_inputfile_format nifti --congeal_inputfiles_list ../input/sample/allfiles --congeal_optimize_algorithm randomwalk --congeal_optimize_randomwalk_kernel 0.1 --congeal_optimize__randomwalk__steps 10 --congeal_optimize__randomwalk__directions 20 --congeal_optimize_error parzen --congeal_error__parzen__sigma 30 --congeal_error__parzen__apriori 1e-06 --congeal_output_prefix ../output/congeal/ --congeal_output_colors_mid 128 --congeal_output_colors_range 256 --congeal_output_sourcegrid 9 --congeal_optimize_progresspoints 4 --congeal_output_average_width 512 --congeal_output_average_height 512 --congeal_initialsteps_translate 0.2 --congeal_initialsteps_rotate 30 --congeal_initialsteps_scale 0.2 --congeal_initialsteps_warp 0.15 --congeal_schedule__n__cache true,true,true,true,true --congeal_schedule__n__downsample 0,0,0,0,0 --congeal_schedule__n__optimize_affine true,false,false,false,false --congeal_schedule__n__warpfield__0__size 1,4,4,4,4 --congeal_schedule__n__warpfield__1__size 1,1,8,8,8 --congeal_schedule__n__warpfield__2__size 1,1,1,16,16 --congeal_schedule__n__warpfield__3__size 1,1,1,1,32 --congeal_schedule__n__optimize_warp__0__ false,true,false,false,false --congeal_schedule__n__optimize_warp__1__ false,false,true,false,false --congeal_schedule__n__optimize_warp__2__ false,false,false,true,false --congeal_schedule__n__optimize_warp__3__ false,false,false,false,true --congeal_schedule__n__optimize_iterations 30,30,30,30,30 --congeal_schedule__n__optimize_samples 50000,50000,500000,500000,500000 --congeal_optimize_bestpoints 1000 --test congeal Configuration file written to /tmp/congealing.config
and the output is the same for both (thanks to the default values, they also equal a press on the Apply button in the GUI without any changes to the input fields):
$ cat /var/tmp/tmp.0.YQNbrV
# experimental
congeal.optimize.bestpoints 1000
test congeal
# input
congeal.inputfiles 30
congeal.inputfile.format nifti
congeal.inputfiles.list ../input/sample/allfiles
# optimization
congeal.optimize.algorithm randomwalk
# randomwalk
congeal.optimize[randomwalk].kernel 0.1
congeal.optimize[randomwalk].steps 10
congeal.optimize[randomwalk].directions 20
# error function
congeal.optimize.error parzen
# parzen error function
congeal.error[parzen].sigma 30
congeal.error[parzen].apriori 1e-06
# output
congeal.output.prefix ../output/congeal/
congeal.output.colors.mid 128
congeal.output.colors.range 256
congeal.output.sourcegrid 9
congeal.optimize.progresspoints 4
congeal.output.average.width 512
congeal.output.average.height 512
# initial steps
congeal.initialsteps.translate 0.2
congeal.initialsteps.rotate 30
congeal.initialsteps.scale 0.2
congeal.initialsteps.warp 0.15
# schedules
n -1
congeal.schedule[{++n}].cache true
congeal.schedule[{$n}].downsample 0
congeal.schedule[{$n}].optimize.affine true
congeal.schedule[{$n}].warpfield[0].size 1
congeal.schedule[{$n}].warpfield[1].size 1
congeal.schedule[{$n}].warpfield[2].size 1
congeal.schedule[{$n}].warpfield[3].size 1
congeal.schedule[{$n}].optimize.warp[0] false
congeal.schedule[{$n}].optimize.warp[1] false
congeal.schedule[{$n}].optimize.warp[2] false
congeal.schedule[{$n}].optimize.warp[3] false
congeal.schedule[{$n}].optimize.iterations 30
congeal.schedule[{$n}].optimize.samples 50000
congeal.schedule[{++n}].cache true
congeal.schedule[{$n}].downsample 0
congeal.schedule[{$n}].optimize.affine false
congeal.schedule[{$n}].warpfield[0].size 4
congeal.schedule[{$n}].warpfield[1].size 1
congeal.schedule[{$n}].warpfield[2].size 1
congeal.schedule[{$n}].warpfield[3].size 1
congeal.schedule[{$n}].optimize.warp[0] true
congeal.schedule[{$n}].optimize.warp[1] false
congeal.schedule[{$n}].optimize.warp[2] false
congeal.schedule[{$n}].optimize.warp[3] false
congeal.schedule[{$n}].optimize.iterations 30
congeal.schedule[{$n}].optimize.samples 50000
congeal.schedule[{++n}].cache true
congeal.schedule[{$n}].downsample 0
congeal.schedule[{$n}].optimize.affine false
congeal.schedule[{$n}].warpfield[0].size 4
congeal.schedule[{$n}].warpfield[1].size 8
congeal.schedule[{$n}].warpfield[2].size 1
congeal.schedule[{$n}].warpfield[3].size 1
congeal.schedule[{$n}].optimize.warp[0] false
congeal.schedule[{$n}].optimize.warp[1] true
congeal.schedule[{$n}].optimize.warp[2] false
congeal.schedule[{$n}].optimize.warp[3] false
congeal.schedule[{$n}].optimize.iterations 30
congeal.schedule[{$n}].optimize.samples 500000
congeal.schedule[{++n}].cache true
congeal.schedule[{$n}].downsample 0
congeal.schedule[{$n}].optimize.affine false
congeal.schedule[{$n}].warpfield[0].size 4
congeal.schedule[{$n}].warpfield[1].size 8
congeal.schedule[{$n}].warpfield[2].size 16
congeal.schedule[{$n}].warpfield[3].size 1
congeal.schedule[{$n}].optimize.warp[0] false
congeal.schedule[{$n}].optimize.warp[1] false
congeal.schedule[{$n}].optimize.warp[2] true
congeal.schedule[{$n}].optimize.warp[3] false
congeal.schedule[{$n}].optimize.iterations 30
congeal.schedule[{$n}].optimize.samples 500000
congeal.schedule[{++n}].cache true
congeal.schedule[{$n}].downsample 0
congeal.schedule[{$n}].optimize.affine false
congeal.schedule[{$n}].warpfield[0].size 4
congeal.schedule[{$n}].warpfield[1].size 8
congeal.schedule[{$n}].warpfield[2].size 16
congeal.schedule[{$n}].warpfield[3].size 32
congeal.schedule[{$n}].optimize.warp[0] false
congeal.schedule[{$n}].optimize.warp[1] false
congeal.schedule[{$n}].optimize.warp[2] false
congeal.schedule[{$n}].optimize.warp[3] true
congeal.schedule[{$n}].optimize.iterations 30
congeal.schedule[{$n}].optimize.samples 500000
congeal.schedules {++n}
Using --launch
When adding a --launch PATH_TO_CONGEAL_EXEC, the congeal executable gets launched rather than printing the path to the generated configuration file.
For example
$ ./CongealingCLI --launch congeal
starts the congeal executable with the configuration file shown above.