Difference between revisions of "Modules:AtlasCreator"
| Line 34: | Line 34: | ||
* Cluster Computation Mode | * Cluster Computation Mode | ||
* using existing Transforms and skipping the Registration (f.e. to re-run a previous generation) | * using existing Transforms and skipping the Registration (f.e. to re-run a previous generation) | ||
| + | |||
| + | ''Please cite this work as:'' | ||
| + | L. Zöllei, M. Shenton, W.M. Wells III, K.M. Pohl. The Impact of Atlas Formation Methods on Atlas-Guided Brain Segmentation, Statistical Registration. In ''Pair-wise and Group-wise Alignment and Atlas Formation Workshop at MICCAI 2007: Tenth International Conference on Medical Image Computing and Computer-Assisted Intervention'', pp. 39 - 46, Brisbane, Australia, 2007 | ||
== Usage == | == Usage == | ||
Revision as of 01:18, 15 April 2011
Home < Modules:AtlasCreatorReturn to Slicer 3.6 Documentation
AtlasCreator
General Information
Module Type & Category
Type: Built-in Loadable Module
Category: Registration
Authors, Collaborators & Contact
- Daniel Haehn, University of Pennsylvania
- Kilian Pohl, University of Pennsylvania
- Contact: Daniel Haehn (haehn@bwh.harvard.edu)
Module Description
The Atlas Creator module aligns images paired with segmentations to generate statistical atlases for several segmented structures.
Features:
- Support for BRAINSFit/CMTK/Congeal toolkits for Registration and Resampling
- Fixed Registration against a template or Dynamic Registration against a mean image
- Normalization of output atlases to a given value
- auto-detection of Structures (labels)
- different Output Casts
- Principal Component Analysis
- Cluster Computation Mode
- using existing Transforms and skipping the Registration (f.e. to re-run a previous generation)
Please cite this work as: L. Zöllei, M. Shenton, W.M. Wells III, K.M. Pohl. The Impact of Atlas Formation Methods on Atlas-Guided Brain Segmentation, Statistical Registration. In Pair-wise and Group-wise Alignment and Atlas Formation Workshop at MICCAI 2007: Tenth International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 39 - 46, Brisbane, Australia, 2007
Usage
This module supports different usage types and interfaces:
- Simple Atlas Creation
- The extended graphical user interface
- The command line interface
- External invocation using the Atlas Creator MRML Node
Use Cases, Examples
Input Data Requirements
The Atlas Creator expects input data to be structured the following way:
- In general each original image is accompanied by a manual segmentation.
- The images and the segmentations have to be in two different folders but have matching filenames.
- All images and segmentations have to be in Slicer-readable format.
- For Example:
- ./originals/case1.nrrd
- ./originals/case2.nrrd
- ./originals/case3.nrrd
- ./segmentations/case1.nrrd
- ./segmentations/case2.nrrd
- ./segmentations/case3.nrrd
| HowTo: Simple Atlas Creation |
| The following steps perform a Pair Fixed Registration against an automatic chosen template for automatically detected structures. |
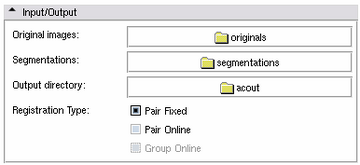
| 1. Select the directories containing the Original Images and the Segmentations in the Input/Output panel. |
| 2. Select an Output Directory in the Input/Output panel. It makes sense to create a new directory to use for the Output. |
| 3. Hit Start! |
| 4. After some wait (minutes or hours!, depending on the number of input cases), the generated atlases and the used template will be loaded into 3D Slicer. |
Output Directory Structure
The Output Directory contains the following content after the Atlas Creator finished.
- ./template.nrrd - The fixed or dynamic template
- ./normalizedIntensityMapOfAligned.nrrd - An intensity map of all aligned cases, divided by the number of cases.
- ./registered/ - A directory containing the registered Original Images
- ./resampled/ - A directory containing the resampled Segmentations used for the Atlas Creation
- ./transforms/ - A directory containing the generated transforms as a result of the registration
- ./atlasX.nrrd - Atlas for label X
- ./atlasY.nrrd - Atlas for label Y
- ./atlasZ.nrrd - Atlas for label Z
- ... more atlases depending on which structures where used
Quick Tour of Features and Use
The Extended Graphical User Interface
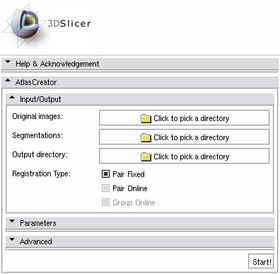
The Atlas Creator module is organized in panels from which only the Input/Output panel is expanded by default. Additional features are provided through an extended interface. This extended interface is divided into the Parameters panel and the Advanced panels which are collapsed by default.
Input/Output Panel
- Original Images:
- Segmentations:
- Output directory:
- Registration Type:
- Pair Fixed:
- Pair Online:
- Group Online:
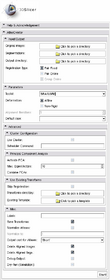
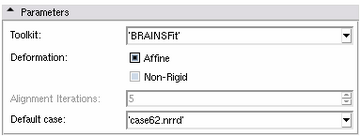
Parameters Panel
- Toolkit:
- Deformation:
- Affine:
- Non-rigid:
- Alignment iterations, for Pair Online:
- Default case, for Pair Fixed:
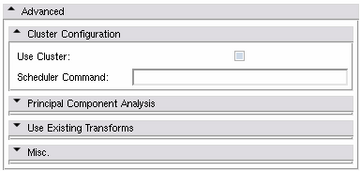
Advanced Panels
Cluster Configuration Panel
The Atlas Creator supports distributing all computations among a Grid Environment.
- Use Cluster:
- Scheduler Command:
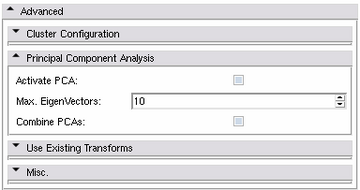
Principal Component Analysis Panel
- Activate PCA:
- Max. Eigenvectors:
- Combine PCAs:
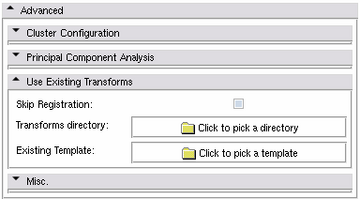
Use Existing Transforms Panel
- Skip Registration:
- Transforms directory:
- Existing Template::
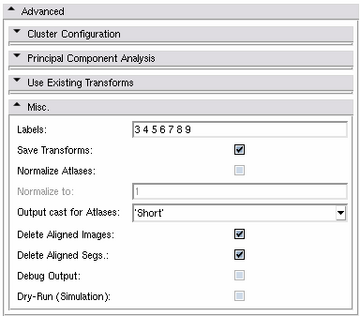
Misc. Panel
- Labels:
- Save Transforms:
- Normalize Atlases:
- Normalize to:
- Output cast for Atlases:
- Delete aligned Images:
- Delete aligned Segs.:
- Debug Output:
- Dry-Run (Simulaton):
The Command Line Interface
Beside using the graphical user interface in 3D Slicer, the Atlas Creator can be accessed using the commandline interface.
$ cd Slicer3-release/lib/Slicer3/Modules/AtlasCreator/atlascreator.py
$ python atlascreator.py --help
AtlasCreator for 3D Slicer
Version v0.4
Usage:
-h, --help
Show this information.
-i, --images DIR
Directory containing original images.
-s, --segmentations DIR
Directory containing segmentations.
-o, --output DIR
Output directory.
[....]
The full documentation of the commandline interface is available on a separate page.
External invocation using the Atlas Creator MRML Node
Development
Notes from the Developer(s)
Separate notes on the development are available on a separate page.
Dependencies
At least BRAINSFit, CMTK or Congeal have to be installed. BRAINSFit is deployed via Slicer, VMTK and Congeal are available as Slicer extensions.
Tests
On the Dashboard, these tests verify that the module is working on various platforms:
- vtkMRMLAtlasCreatorNodeTest1 Modules/AtlasCreator/Cxx/Testing/vtkMRMLAtlasCreatorNodeTest1.cxx
- AtlasCreatorLaunchFixedTest Modules/AtlasCreator/Cxx/Testing/vtkMRMLAtlasCreatorNodeLaunchTest1.py
- AtlasCreatorLaunchFixedFailProofTest Modules/AtlasCreator/Cxx/Testing/vtkMRMLAtlasCreatorNodeLaunchTest2.py
- AtlasCreatorLaunchDynamicTest Modules/AtlasCreator/Cxx/Testing/vtkMRMLAtlasCreatorNodeLaunchTest3.py
Known bugs
Usability issues
- Congeal support is still Under Construction.
Source code & documentation
Source code:
- Available in the 3D Slicer Trunk: Modules/AtlasCreator
Doxygen documentation:
More Information
Acknowledgment
The research was funded by an ARRA supplement to NIH NCRR (P41 RR13218).
The Atlas Creator module benefits from several different Toolkits: Thank you for BRAINSFit (Hans Johnson et al.), for CMTK (Torsten Rolfing et al.) and for Congeal (J. De Bonet, L. Zöllei and W.M. Wells III).