Main Page/SlicerCommunity/2020
Go to 2022 :: 2021 :: 2020 :: 2019 :: 2018 :: 2017 :: 2016 :: 2015 :: 2014-2011 :: 2010-2000
The community that relies on 3D Slicer is large and active: (numbers below updated on December 1st, 2023)
- 1,467,466+ downloads in the last 11 years (269,677 in 2023, 206,541 in 2022)
- over 17.900+ literature search results on Google Scholar
- 2,147+ papers on PubMed citing the Slicer platform paper
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J-C., Pujol S., Bauer C., Jennings D., Fennessy F.M., Sonka M., Buatti J., Aylward S.R., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magnetic Resonance Imaging. 2012 Nov;30(9):1323-41. PMID: 22770690. PMCID: PMC3466397.
- 39 events in open source hackathon series continuously running since 2005 with 3260 total participants
- Slicer Forum with +8,138 subscribers has approximately 275 posts every week
The following is a sample of the research performed using 3D Slicer outside of the group that develops it. in 2020
We monitor PubMed and related databases to update these lists, but if you know of other research related to the Slicer community that should be included here please email: marianna (at) bwh.harvard.edu.
Contents
- 1 2020
- 1.1 Quantitative Chest CT Analysis in COVID-19 to Predict the Need for Oxygenation Support and Intubation
- 1.2 3D Reconstruction of Wilms' Tumor and Kidneys in Children: Variability, Usefulness and Constraints
- 1.3 Digital Three-Dimensional Visualization of Intrabony Periodontal Defects for Regenerative Surgical Treatment Planning
- 1.4 Lung CT Segmentation to Identify Consolidations and Ground Glass Areas for Quantitative Assesment of SARS-CoV Pneumonia
- 1.5 Feasibility of MRI Radiomics for Predicting KRAS Mutation in Rectal Cancer
- 1.6 Using Individualized Three-Dimensional Printed Airway Models to Guide Airway Stent Implantation
- 1.7 3D Slicer and Sina Appilication for Surgical Planning of Giant Invasive Spinal Schwannoma With Scoliosis: A Case Report and Literature Review
- 1.8 A Novel CNN Algorithm for Pathological Complete Response Prediction Using an I-SPY TRIAL Breast MRI Database
- 1.9 The Effect of Registration on Voxel-Wise Tofts Model Parameters and Uncertainties from DCE-MRI of Early-Stage Breast Cancer Patients Using 3DSlicer
- 1.10 3D Slicer Craniomaxillofacial Modules Support Patient-Specific Decision-Making for Personalized Healthcare in Dental Research
- 1.11 GeodesicSlicer: a Slicer Toolbox for Targeting Brain Stimulation
- 1.12 A Novel Open-Source Software-Based High-Precision Workflow for Target Definition in Cardiac Radioablation
- 1.13 A Novel Method for Observing Tumor Margin in Hepatoblastoma Based on Microstructure 3D Reconstruction
- 1.14 Setting Up 3D Printing Services for Orthopaedic Applications: A Step-by-Step Guide and an Overview of 3DBioSphere
- 1.15 SlicerArduino: A Bridge between Medical Imaging Platform and Microcontroller
- 1.16 Quantification of Nasal Septal Deviation With Computed Tomography Data
- 1.17 CroSSED Sequence, a New Tool for 3D Processing in Geosciences Using the Free Software 3DSlicer
- 1.18 Anatomical Investigation of Middle Mesial Canals of Mandibular Molars in a Middle Eastern Population: A Cross-sectional Cone-beam Computed Tomography Study
- 1.19 Radiomic Features of Primary Tumor by Lung Cancer Stage: Analysis in BRAF Mutated Non-Small Cell Lung Cancer
- 1.20 New Classification of Cochlear Hypoplasia Type Malformation: Relevance in Cochlear Implantation
- 1.21 Assessment of the Severity of Coronavirus Disease: Quantitative Computed Tomography Parameters versus Semiquantitative Visual Score
- 1.22 Timing of Ossification of the Anterior Skull Base in Syndromic Synostosis
- 1.23 Three-Dimensional Volumetric Changes in Posterior Vault Distraction With Distraction Osteogenesis
- 1.24 Evaluation of Imaging Software Accuracy for 3-Dimensional Analysis of the Mandibular Condyle. A Comparative Study Using a Surface-to-Surface Matching Technique
- 1.25 Temporomandibular Joint Damage in K/BxN Arthritic Mice
- 1.26 Synchrotron Radiation-Based Reconstruction of the Human Spiral Ganglion: Implications for Cochlear Implantation
- 1.27 Visualization of Mucosal Field in HPV Positive and Negative Oropharyngeal Squamous Cell Carcinomas: Combined Genomic and Radiology Based 3D Model
- 1.28 Using of a Dismountable 3D-model of the Collecting System with Color Segmentation to Improve the Learning Curve of Residents
- 1.29 Manual and Semiautomatic Segmentation of Bone Sarcomas on MRI Have High Similarity
- 1.30 Measurement Error and Reliability of Three Available 3D Superimposition Methods in Growing Patients
- 1.31 Prostate Multiparametric Magnetic Resonance Imaging Features Following Partial Gland Cryoablation
- 1.32 3D Printing Method for Next-Day Acetabular Fracture Surgery Using a Surface Filtering Pipeline: Feasibility and 1-Year Clinical Results
- 1.33 Convolutional Neural Network-based MR Image Analysis for Alzheimer's Disease Classification
2020
Quantitative Chest CT Analysis in COVID-19 to Predict the Need for Oxygenation Support and Intubation
|
Publication: Eur Radiol. 2020 Dec;30(12):6770-8. PMID: 32591888 | PDF
Institution: Department of Diagnostic and Interventional Radiology, Humanitas Clinical and Research Center - IRCCS, Milan, Italy. Abstract: We performed a single-centre retrospective study on COVID-19 patients hospitalised from January 25, 2020, to April 28, 2020, who received CT at admission prompted by respiratory symptoms such as dyspnea or desaturation. QCT was performed using a semi-automated method (3D Slicer). Lungs were divided by Hounsfield unit intervals. Compromised lung (%CL) volume was the sum of poorly and non-aerated volumes (- 500, 100 HU). We collected patient's clinical data including oxygenation support throughout hospitalisation. |
3D Reconstruction of Wilms' Tumor and Kidneys in Children: Variability, Usefulness and Constraints
|
Publication: J Pediatr Urol. 2020 Dec;16(6):830.e1-830.e8. PMID: 32893166 Authors: Chaussy Y, Vieille L, Lacroix E, Lenoir M, Marie F, Corbat L, Henriet J, Auber F. Institution: Department of Pediatric Surgery, CHU Besançon, Besançon, France. Abstract: 14 scans from 12 patients were manually or semi-automatically segmented by 2 teams using 3D Slicer software. Inter-individual variability of 3D reconstructions was measured based on the Dice index. The utility of 3D reconstructions for the surgical planning was evaluated by 4 pediatric surgeons using a 5-point Likert scale. The possibility of undertaking NSS was evaluated according to the criteria defined in the Umbrella SIOP-RTSG 2016 protocol. |
Digital Three-Dimensional Visualization of Intrabony Periodontal Defects for Regenerative Surgical Treatment Planning
|
Publication: BMC Oral Health. 2020 Dec 1;20(1):351. PMID: 33261592 | PDF Authors: Palkovics D, Mangano FG, Nagy K, Windisch P. Institution: Department of Periodontology, Semmelweis University, Budapest, Hungary. Abstract: Four patients with a total of six intrabony periodontal defects were enrolled in the present study. Two months following initial periodontal treatment, a CBCT scan was taken. The novel semi-automatic segmentation method was performed in an open-source medical image processing software, 3D Slicer to acquire virtual 3D models of alveolar and dental structures. Intrasurgical and digital measurements were taken, and results were compared to validate the accuracy of the digital models. Defect characteristics were determined prior to surgery with conventional diagnostic methods and 3D virtual models. Diagnostic assessments were compared to the actual defect morphology during surgery. |
Lung CT Segmentation to Identify Consolidations and Ground Glass Areas for Quantitative Assesment of SARS-CoV Pneumonia
|
Publication: J Vis Exp. 2020 Dec 19;(166). PMID: 33393515 Authors: Cattabriga A, Cocozza MA, Vara G, Coppola F, Golfieri R. Institution: Department of Diagnostic and Specialty Medicine, Policlinico Sant'Orsola-Malpighi, University of Bologna, Bologna, Italy. Abstract: Segmentation is a complex task, faced by radiologists and researchers as radiomics and machine learning grow in potentiality. The process can either be automatic, semi-automatic, or manual, the first often not being sufficiently precise or easily reproducible, and the last being excessively time consuming when involving large districts with high-resolution acquisitions. A high-resolution CT of the chest is composed of hundreds of images, and this makes the manual approach excessively time consuming. Furthermore, the parenchymal alterations require an expert evaluation to be discerned from the normal appearance; thus, a semi-automatic approach to the segmentation process is, to the best of our knowledge, the most suitable when segmenting pneumonias, especially when their features are still unknown. For the studies conducted in our institute on the imaging of COVID-19, we adopted 3D Slicer, a freeware software produced by the Harvard University, and combined the threshold with the paint brush instruments to achieve fast and precise segmentation of aerated lung, ground glass opacities, and consolidations. When facing complex cases, this method still requires a considerable amount of time for proper manual adjustments, but provides an extremely efficient mean to define segments to use for further analysis, such as the calculation of the percentage of the affected lung parenchyma or texture analysis of the ground glass areas. |
Feasibility of MRI Radiomics for Predicting KRAS Mutation in Rectal Cancer
|
Publication: Curr Med Sci. 2020 Dec;40(6):1156-60. PMID: 33428144 Authors: Guo XF, Yang WQ, Yang Q, Yuan ZL, Liu YL, Niu XH, Xu HB. Institution: Department of Radiology, Zhongnan Hospital of Wuhan University, Wuhan, China. Abstract: The mutation status of KRAS is a significant biomarker in the prognosis of rectal cancer. This study investigated the feasibility of MRI-based radiomics in predicting the mutation status of KRAS with a composite index which could be an important criterion for KRAS mutation in clinical practice. In this retrospective study, a total of 127 patients with rectal cancer were enrolled. 3D Slicer was used to extract the radiomics features from the MRI images, and sparse support vector machine (SVM) with linear kernel was applied for feature reduction. The radiomics classifier for predicting the KRAS status was then constructed by Linear Discriminant Analysis (LDA) and its performance was evaluated. The composite index was determined with LDA model. Out of 127 rectal cancer subjects, there were 44 KRAS mutation cases and 83 wild cases. A total of 104 radiomics features were extracted, 54 features were filtered by linear SVM with L1-norm regularization and 6 features that had no significant correlations within them were finally selected. The radiomics classifier constructed using the 6 features featured an AUC value of 0.669 (specificity: 0.506; sensitivity: 0.773) with LDA. Furthermore, the composite index (Radscore) had statistically significant difference between the KRAS mutation and wild groups. It is suggested that the MRI-based radiomics has the potential in predicting the KRAS status in patients with rectal cancer, which may enhance the diagnostic value of MRI in rectal cancer. |
Using Individualized Three-Dimensional Printed Airway Models to Guide Airway Stent Implantation
|
Publication: Interact Cardiovasc Thorac Surg. 2020 Dec 7;31(6):900-3. PMID: 33150423 Authors: Xu J, Sullivan C, Ong HX, Williamson JP, Traini D, Hersch N, Byrom M, Young PM. Institution: Respiratory Technology Group, Woolcock Institute of Medical Research, Sydney, NSW, Australia. Abstract: Airway stents are used to manage central airway obstructions by restoring airway patency. Current manufactured stents are limited in shape and size, which pose issues in stent fenestrations needed to be manually created to allow collateral ventilation to airway branches. The precise location to place these fenestrations can be difficult to predict based on 2-dimensional computed tomography images. Inspiratory computed tomography scans were obtained from 3 patients and analysed using 3D Slicer™, Blender™ and AutoDesk® Meshmixer™ programmes to obtain working 3D-airway models, which were 3D printed. Stent customizations were made based on 3D-model dimensions, and fenestrations into the stent were cut. The modified stents were then inserted as per usual technique. Two patients reported improved airway performance; however, stents were later removed due to symptoms related to in-stent sputum retention. In a third patient, the stent was removed a few weeks later due to the persistence of fistula leakage. The use of a 3D-printed personalized airway model allowed for more precise stent customization, optimizing stent fit and allowing for cross-ventilation of branching airways. We determine that an airway model is a beneficial tool for stent optimization but does not prevent the development of some stent-related complications such as airway secretions. |
3D Slicer and Sina Appilication for Surgical Planning of Giant Invasive Spinal Schwannoma With Scoliosis: A Case Report and Literature Review
|
Publication: Neurochirurgie. 2020 Nov;66(5):396-399. PMID: 32777234 Authors: Hou X, Yang DD, Li D, Zeng L, Li C. Institution: School of Clinical Medicine, Chengdu University of TCM, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China. Abstract: The surgical management of giant invasive spinal schwannomas (GISSs) has been discussed previously, and most cases are treated via the posterior approach. However, when the tumor grows beyond the spine and greatly pushes the surrounding tissues, the combined anterior-posterior approach may be a better choice. However, the anterior approach can be challenging when there is a lack of knowledge regarding the surrounding abdominal structures. Case description: A 67-year-old male suffered from slight scoliosis and an unstable spine due to GISS, which led to a long history of lower back pain and abnormal gait. Here, we report a novel method combining the use of 3D Slicer and the Sina application to help surgeons locate such lesions and better understand the three-dimensional (3D) relationship between the tumor and surrounding tissues. The proposed method promotes complete excision, shortens operation time, and reduces related complications. Conclusion: We recommend using the combination of 3D Slicer and Sina to assist surgeons in handling accurate 3D information of GISS while simultaneously simulating the surgery. |
A Novel CNN Algorithm for Pathological Complete Response Prediction Using an I-SPY TRIAL Breast MRI Database
|
Publication: Magn Reson Imaging. 2020 Nov;73:148-51. PMID: 32889091 | PDF Authors: Liu MZ, Mutasa S, Chang P, Siddique M, Jambawalikar S, Ha R. Institution: Department of Medical Physics, Columbia University Medical Center, New York, NY, USA. Abstract: From the I-SPY TRIAL breast MRI database, 131 patients from 9 institutions were successfully downloaded for analysis. First post-contrast MRI images were used for 3D segmentation using 3D Slicer. Our CNN was implemented entirely of 3 × 3 convolutional kernels and linear layers. The convolutional kernels consisted of 6 residual layers, totaling 12 convolutional layers. Dropout with a 0.5 keep probability and L2 normalization was utilized. Training was implemented by using the Adam optimizer. A 5-fold cross validation was used for performance evaluation. Software code was written in Python using the TensorFlow module on a Linux workstation with one NVidia Titan X GPU. Funding: R38 CA231577/CA/NCI NIH HHS/United States |
The Effect of Registration on Voxel-Wise Tofts Model Parameters and Uncertainties from DCE-MRI of Early-Stage Breast Cancer Patients Using 3DSlicer
|
Publication: J Digit Imaging. 2020 Oct;33(5):1065-72. PMID: 32748300 Authors: Mouawad M, Biernaski H, Brackstone M, Lock M, Kornecki A, Shmuilovich O, Ben-Nachum I, Prato FS, Thompson RT, Gaede S, Gelman N. Institution: Medical Biophysics, Western University, London, ON, Canada. Abstract: We quantitatively investigate the influence of image registration, using open-source software (3DSlicer), on kinetic analysis (Tofts model) of dynamic contrast enhanced MRI of early-stage breast cancer patients. We also show that registration computation time can be reduced by reducing the percent sampling (PS) of voxels used for estimation of the cost function. DCE-MRI breast images were acquired on a 3T-PET/MRI system in 13 patients with early-stage breast cancer who were scanned in a prone radiotherapy position. Images were registered using a BSpline transformation with a 2 cm isotropic grid at 100, 20, 5, 1, and 0.5PS (BRAINSFit in 3D Slicer). Signal enhancement curves were analyzed voxel-by-voxel using the Tofts kinetic model. Comparing unregistered with registered groups, we found a significant change in the 90th percentile of the voxel-wise distribution of Ktrans. We also found a significant reduction in the following: (1) in the standard error (uncertainty) of the parameter value estimation, (2) the number of voxel fits providing unphysical values for the extracellular-extravascular volume fraction (ve > 1), and (3) goodness of fit. We found no significant differences in the median of parameter value distributions (Ktrans, ve) between unregistered and registered images. Differences between parameters and uncertainties obtained using 100PS versus 20PS were small and statistically insignificant. As such, computation time can be reduced by a factor of 2, on average, by using 20PS while not affecting the kinetic fit. The methods outlined here are important for studies including a large number of post-contrast images or number of patient images. |
3D Slicer Craniomaxillofacial Modules Support Patient-Specific Decision-Making for Personalized Healthcare in Dental Research
|
Publication: Multimodal Learn Clin Decis Support Clin Image Based Proc (2020). 2020 Oct;12445:44-53. PMID: 33415323 | PDF Authors: Bianchi J, Paniagua B, De Oliveira Ruellas AC, Fillion-Robin JC, Prietro JC, Gonçalves JR, Hoctor J, Yatabe M, Styner M, Li T, Gurgel ML, Chaves CM, Massaro C, Garib DG, Vilanova L, Castanha Henriques JF, Aliaga-Del Castillo A, Janson G, Iwasaki LR, Nickel JC, Evangelista K, Cevidanes L. Institution: University of Michigan, 1011 North University Ave, Ann Arbor, USA. Abstract: The biggest challenge to improve the diagnosis and therapies of Craniomaxillofacial conditions is to translate algorithms and software developments towards the creation of holistic patient models. A complete picture of the individual patient for treatment planning and personalized healthcare requires a compilation of clinician-friendly algorithms to provide minimally invasive diagnostic techniques with multimodal image integration and analysis. We describe here the implementation of the open-source Craniomaxillofacial module of the 3D Slicer software, as well as its clinical applications. This paper proposes data management approaches for multisource data extraction, registration, visualization, and quantification. These applications integrate medical images with clinical and biological data analytics, user studies, and other heterogeneous data. Funding: R01 DE024450/DE/NIDCR NIH HHS/United States |
GeodesicSlicer: a Slicer Toolbox for Targeting Brain Stimulation
|
Publication: Neuroinformatics. 2020 Oct;18(4):509-516. PMID: 32125609 Authors: Briend F, Leroux E, Nathou C, Delcroix N, Dollfus S, Etard O. Institution: Normandie University, Caen, France. Abstract: NonInvasive Brain Stimulation (NIBS) is a potential therapeutic tool with growing interest, but neuronavigation-guided software and tools available for the target determination are mostly either expensive or closed proprietary applications. To address these limitations, we propose GeodesicSlicer, a customizable, free, and open-source NIBS therapy research toolkit. GeodesicSlicer is implemented as an extension for the widely used 3D Slicer medical image visualization and analysis application platform. GeodesicSlicer uses cortical stimulation target from either functional or anatomical images to provide functionality specifically designed for NIBS therapy research. The provided algorithms are tested and they are accessible through a convenient graphical user interface. Modules have been created for NIBS target determination according to the position of the electrodes in the 10-20 system electroencephalogram and calculating correction factors to adjust the repetitive Transcranial Magnetic Stimulation (rTMS) dose for the treatment. Two illustrative examples are processing with the module. This new open-source software has been developed for NIBS therapy: GeodesicSlicer is an alternative for laboratories that do not have access to neuronavigation system. The triangulation-based MRI-guided method presented here provides a reproducible and inexpensive way to position the TMS coil that may be used without the use of a neuronavigation system. |
A Novel Open-Source Software-Based High-Precision Workflow for Target Definition in Cardiac Radioablation
|
Publication: J Cardiovasc Electrophysiol. 2020 Oct;31(10):2689-95. PMID: 32648343 Authors: Hohmann S, Henkenberens C, Zormpas C, Christiansen H, Bauersachs J, Duncker D, Veltmann C. Institution: Hannover Heart Rhythm Center, Department of Cardiology and Angiology, Hannover Medical School, Hannover, Germany. Abstract: Using the free open-source 3D Slicer software platform we established a workflow for high-precision target definition based on electroanatomic maps. An import plug-in for 3D Slicer has been designed that reads electroanatomic maps generated with three mapping systems in widespread clinical use. Using our proposed workflow in a real-world patient case we were able to align the map to the computed tomography (CT) with a mean distance of 3.1 mm. Thus, points defined on the map were translated into CT space with high accuracy and a radiotherapy treatment volume was defined in CT space based on these map-derived points. |
A Novel Method for Observing Tumor Margin in Hepatoblastoma Based on Microstructure 3D Reconstruction
|
Publication: Fetal Pediatr Pathol. 2020 Sep 24:1-10. PMID: 32969743 Authors: Liu J, Wu X, Xu C, Ma M, Zhao J, Li M, Yu Q, Hao X, Wang G, Wei B, Xia N, Dong Q. Institution: Department of Pediatric Surgery, Affiliated Hospital of Qingdao University, Qingdao University, China. Abstract: We investigated three-dimensional (3 D) reconstruction for the assessment of the tumor margin microstructure of hepatoblastoma (HB). Methods: Eleven surgical resections of childhood hepatoblastomas obtained between September 2018 and December 2019 were formalin-fixed, paraffin-embedded, serially sectioned at 4 μm, stained with hematoxylin and eosin (every 19th and 20th section stained with alpha-fetoprotein and glypican 3), and the digital images of all sections were acquired at 100× followed by image registration using the B-spline based method with modified residual complexity. Reconstruction was performed using 3D Slicer software. Results: The reconstructed orthogonal 3 D images clearly presented the internal microstructure of the tumor margin. The rendered 3 D image could be rotated at any angle. Conclusions: Microstructure 3 D reconstruction is feasible for observing the pathological structure of the HB tumor margin. |
Setting Up 3D Printing Services for Orthopaedic Applications: A Step-by-Step Guide and an Overview of 3DBioSphere
|
Publication: Indian J Orthop. 2020 Sep 15;54(Suppl 2):217-27. PMID: 33194095 Authors: Shah D, Naik L, Paunipagar B, Rasalkar D, Chaudhary K, Bagaria V. Institution: Department of Orthopaedics, Sir HN Reliance Foundation Hospital, Mumbai, India. Abstract: Introduction: 3D printing has widespread applications in orthopaedics including creating biomodels, patient-specific instruments, implants, and developing bioprints. 3DGraphy or printing 3D models enable the surgeon to understand, plan, and simulate different procedures on it. Despite widespread applications in non-healthcare specialties, it has failed to gain traction in healthcare settings. This is perhaps due to perceived capital expenditure cost and the lack of knowledge and skill required to execute the process. Purpose: This article is written with an aim to provide step-by-step instructions for setting up a cost-efficient 3D printing laboratory in an institution or standalone radiology centre. The article with the help of video modules will explain the key process of segmentation, especially the technique of edge detection and thresholding which are the heart of 3D printing. Conclusion: This is likely to enable the practising orthopaedician and radiologist to set up a 3D printing unit in their departments or even standalone radiology centres at minimal startup costs. This will enable maximal utilisation of this technology that is likely to bring about a paradigm shift in planning, simulation, and execution of complex surgeries. Keywords: 3D materials; 3D model; 3D printers; 3D printing; 3D Slicer; 3DGraphy; Patient-specific instruments; STL; Segmentation; Thresholding. |
SlicerArduino: A Bridge between Medical Imaging Platform and Microcontroller
|
Publication: Bioengineering (Basel). 2020 Sep 11;7(3):109. PMID: 32932840 | PDF Authors: Zaffino P, Merola A, Leuzzi D, Sabatino V, Cosentino C, Spadea MF. Institution: Department of Clinical and Experimental Medicine, University "Magna Graecia" of Catanzaro, Catanzaro, Italy. Abstract: Interaction between medical image platform and external environment is a desirable feature in several clinical, research, and educational scenarios. In this work, the integration between 3D Slicer package and Arduino board is introduced, enabling a simple and useful communication between the two software/hardware platforms. The open source extension, programmed in Python language, manages the connection process and offers a communication layer accessible from any point of the medical image suite infrastructure. Deep integration with 3D Slicer code environment is provided and a basic input-output mechanism accessible via GUI is also made available. To test the proposed extension, two exemplary use cases were implemented: (1) INPUT data to 3D Slicer, to navigate on basis of data detected by a distance sensor connected to the board, and (2) OUTPUT data from 3D Slicer, to control a servomotor on the basis of data computed through image process procedures. Both goals were achieved and quasi-real-time control was obtained without any lag or freeze, thus boosting the integration between 3D Slicer and Arduino. This integration can be easily obtained through the execution of few lines of Python code. In conclusion, SlicerArduino proved to be suitable for fast prototyping, basic input-output interaction, and educational purposes. The extension is not intended for mission-critical clinical tasks. |
Quantification of Nasal Septal Deviation With Computed Tomography Data
|
Publication: J Craniofac Surg. 2020 Sep;31(6):1659-1663. PMID: 32502103 Authors: Denour E, Roussel LO, Woo AS, Boyajian M, Crozier J. Institution: Department of Molecular Pharmacology, Physiology, and Biotechnology, Brown University, Providence, RI, USA. Abstract: A retrospective study was conducted at a large academic center. One hundred four patients who underwent CT scans of the face were selected from a computer imaging database. Demographic variables were screened to ensure an equal number of men and women in different age groups. Digital Imaging and Communications in Medicine files were imported for 3D nasal cavity segmentation using 3D Slicer software. A volumetric analysis was performed to determine 3D NSD ratios. These values were compared to previously reported methods of obtaining objective 2D NSD measures using OsiriX and MATLAB software. Maximum deviation values were calculated using OsiriX, while the root mean square values were retrieved using MATLAB. Deviation area and curve to line ratios were both quantified using OsiriX and MATLAB. |
CroSSED Sequence, a New Tool for 3D Processing in Geosciences Using the Free Software 3DSlicer
|
Publication: Sci Data. 2020 Aug 14;7(1):270. PMID: 32796857 | PDF Authors: Dorador J, Rodríguez-Tovar FJ. Institution: Department of Earth Sciences, Royal Holloway University of London, Egham, UK. Abstract: The scientific application of 3D imaging has evolved significantly over recent years. These techniques make it possible to study internal features by non-destructive analysis. Despite its potential, the development of 3D imaging in the Geosciences is behind other fields due to the high cost of commercial software and the scarce free alternatives. Most free software was designed for the Health Sciences, and the pre-settled workflows are not suited to geoscientific materials. Thus, an outstanding challenge in the Geosciences is to define workflows using free alternatives for Computed Tomography (CT) data processing, promoting data sharing, reproducibility, and the development of specific extensions. We present CroSSED, a processing sequence for 3D reconstructions of CT data, using 3D Slicer, a popular application in medical imaging. Its usefulness is exemplified in the study of burrows that have low-density contrast with respect to the host sediment. For geoscientists who have access to CT data and wish to reconstruct 3D structures, this method offers a wide range of possibilities and contributes to open-science and applied CT studies. |
Anatomical Investigation of Middle Mesial Canals of Mandibular Molars in a Middle Eastern Population: A Cross-sectional Cone-beam Computed Tomography Study
|
Publication: J Contemp Dent Pract. 2020 Aug 1;21(8):910-5. PMID: 33568614 Authors: Inaty E, Jabre C, Haddad G, Nehme W, Khalil I, Naaman A, Zogheib C. Institution: Department of Endodontics, Saint Joseph University, Faculty of Dentistry, Riad el Solh, Beirut, Lebanon. Abstract: The presence of MMC in the mesial roots of 505 mandibular molars of 200 patients was analyzed using cone-beam computed tomography (CBCT). Then, the position of the MMC orifices with respect to the pulpal floor and the main canals orifices, and the width of dentin along the canal toward the furcation were determined using 3D Slicer v.4.10.1. |
Radiomic Features of Primary Tumor by Lung Cancer Stage: Analysis in BRAF Mutated Non-Small Cell Lung Cancer
|
Publication: Transl Lung Cancer Res. 2020 Aug;9(4):1441-51. PMID: 32953516 | PDF Authors: Padole A, Singh R, Zhang EW, Mendoza DP, Dagogo-Jack I, Kalra MK, Digumarthy SR. Institution: Division of Thoracic Imaging, Massachusetts General Hospital, Boston, MA, USA. Abstract: Our IRB approved study included 62 patients with BRAF mutations (V600 in 27 and non-V600 in 35 patients), who underwent contrast-enhanced chest CT. Tumor stage was determined based on the 8th edition of TNM staging. Two thoracic radiologists assessed the primary tumor imaging features such, including tumor size (maximum and minimum dimensions) and density (Hounsfield units, HU). De-identified transverse CT images (DICOM) were processed with 3D Slicer v.4.7 for manual lesion segmentation and estimation of radiomic features. Descriptive statistics, multivariate logistic regression, and receiver operating characteristics (ROC) were performed. |
New Classification of Cochlear Hypoplasia Type Malformation: Relevance in Cochlear Implantation
|
Publication: J Int Adv Otol. 2020 Aug;16(2):153-7. PMID: 32784151 | PDF Authors: Halawani RT, Dhanasingh A. Institution: Ohud General Hospital, Ministry of Health, AL Medina, Kingdom of Saudi Arabia. Abstract: Preoperative computed-tomography (CT) scans of cochlear implant (CI) candidates (N=31) from various clinics across the world with CH type malformation were taken for analysis. CT dataset were loaded into 3D Slicer freeware for three-dimensional (3D) segmentation of the inner-ear by capturing complete inner-ear structures from the entire dataset. Cochlear size in terms of diameter of available cochlear basal turn and length of cochlear lumen was measured from the dataset. In addition, structural connection between IAC and cochlear portions was scrutinized, which is highly relevant to the proposed CH classification in this study. |
|
Publication: Korean J Radiol. 2020 Aug;21(8):998-1006. PMID: 32677384 | PDF Authors: Yin X, Min X, Nan Y, Feng Z, Li B, Cai W, Xi X, Wang L. Institution: Department of Radiology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. Abstract: We retrospectively enrolled 187 patients with COVID-19 treated at Tongji Hospital of Tongji Medical College from February 15, 2020, to February 29, 2020. Demographic data, imaging characteristics, and clinical data were collected, and based on the clinical classification of severity, patients were divided into groups 1 (mild) and 2 (severe/critical). A semiquantitative visual score was used to estimate the lesion extent. 3D Slicer was used to precisely quantify the volume and CT value of the lung and lesions. Correlation coefficients of the quantitative CT parameters, semiquantitative visual score, and clinical classification were calculated using Spearman's correlation. A receiver operating characteristic curve was used to compare the accuracies of quantitative and semi-quantitative methods. |
Timing of Ossification of the Anterior Skull Base in Syndromic Synostosis
|
Publication: J Craniofac Surg. 2020 Jul-Aug;31(5):1256-1260. PMID: 32282683 Authors: Nassar AH, Mercan E, Massenburg BB, Lee A, Brown J, Skladman R, Guo Y, Hopper RA. Institution: Department of Neurological Surgery, University of Washington, Seattle, WA, USA. Abstract: The anterior skull base undergoes a progressive ossification after birth. This has implications on the epidural dissection of early trans-craniofacial osteotomy procedures such as monobloc advancements. Our purpose was to determine the rate of ossification in syndromic synostosis patients relative to a normal cohort to establish when maturation of the anterior skull base is complete. The authors analyzed CT scans from 35 patients with Crouzon, Apert or Pfeiffer syndrome, and 84 patients without any craniofacial anomaly between the ages of 0 and 6 years. The non-ossified anterior skull base area was measured using 3D Slicer. The authors compared the sizes of the defects at different ages between the three syndromes and with the control group using Mann-Whitney test. Significance was set at P < 0.05. All patients less than 12 months of age had a measurable defect anterior to the cribriform whereas patients greater than five years of age had full ossification of the anterior skull base with no evidence of defect. The relationship of defect size and age at scan was non-linear, with most defects closing rapidly in the first six months. The temporal closure pattern of the defect was similar between the three syndromes and the control group with no significant difference. Our findings indicate that syndromic children undergo skull base maturation at the same rate as non-syndromic cases, with the majority ossified by three years of age. Anterior skull base surgeries performed before three years should optimize visualization of this area during dissection. |
Three-Dimensional Volumetric Changes in Posterior Vault Distraction With Distraction Osteogenesis
|
Publication: J Craniofac Surg. 2020 Jul-Aug;31(5):1301-6. PMID: 32282486 Authors: Thatikunta M, Pearson L, Nguyen C, John K, Abolfotoh M, Mutchnick I, Gump W, Chariker M, Moriarty T, Rapp SJ.
Abstract: From 2007 to 2019, a single institution retrospective review revealed 232 craniosynostosis patients. Fourteen demonstrated lambdoid synostosis (6%), and of those, 11 patients were included in the study due to treatment with PVDO or representative sample. Six patients had unilateral synostosis and 5 had bilateral synostosis. Imaging protocol for PVDO patients included preoperative head CT within 1 month of surgery and 8 weeks following distraction cessation with weekly skull plain films. 3D volumetric analyses were performed on pre and postoperative head CT using 3D Slicer software. |
Evaluation of Imaging Software Accuracy for 3-Dimensional Analysis of the Mandibular Condyle. A Comparative Study Using a Surface-to-Surface Matching Technique
|
Publication: Int J Environ Res Public Health. 2020 Jul 3;17(13):4789. PMID: 32635238 | PDF Authors: Lo Giudice A, Quinzi V, Ronsivalle V, Farronato M, Nicotra C, Indelicato F, Isola G. Institution: Department of General Surgery and Surgical-Medical Specialties, Section of Orthodontics, School of Dentistry, University of Catania, Catania, Italy. Abstract: The aim of this study was to assess the accuracy of 3D rendering of the mandibular condylar region obtained from different semi-automatic segmentation methodology. A total of 10 Cone beam computed tomography (CBCT) were selected to perform semi-automatic segmentation of the condyles by using three free-source software (Invesalius, version 3.0.0, Centro de Tecnologia da Informação Renato Archer, Campinas, SP, Brazil; ITK-Snap, version2.2.0; 3D Slicer, version 4.10.2) and one commercially available software Dolphin 3D (Dolphin Imaging, version 11.0, Chatsworth, CA, USA). The same models were also manually segmented (Mimics, version 17.01, Materialise, Leuven, Belgium) and set as ground truth. The accuracy of semi-automatic segmentation was evaluated by (1) comparing the volume of each semi-automatic 3D rendered condylar model with that obtained with manual segmentation, (2) deviation analysis of each 3D rendered mandibular models with those obtained from manual segmentation. No significant differences were found in the volumetric dimensions of the condylar models among the tested software (p > 0.05). However, the color-coded map showed underestimation of the condylar models obtained with ITK-Snap and Slicer 3D, and overestimation with Dolphin 3D and Invesalius. Excellent reliability was found for both intra-observer and inter-observer readings. Despite the excellent reliability, the present findings suggest that data of condylar morphology obtained with semi-automatic segmentation should be taken with caution when an accurate definition of condylar boundaries is required. |
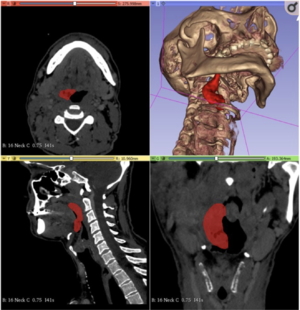
Temporomandibular Joint Damage in K/BxN Arthritic Mice
|
Publication: Int J Oral Sci. 2020 Feb 6;12(1):5. PMID: 32024813 | PDF Authors: Kuchler-Bopp S, Mariotte A, Strub M, Po C, De Cauwer A, Schulz G, Van Bellinghen X, Fioretti F, Clauss F, Georgel P, Benkirane-Jessel N, Bornert F. Institution: INSERM (French National Institute of Health and Medical Research), UMR 1260, Regenerative NanoMedicine (RNM), FMTS, Strasbourg, France. Abstract: Rheumatoid arthritis (RA) is an autoimmune disease affecting 1% of the world population and is characterized by chronic inflammation of the joints sometimes accompanied by extra-articular manifestations. K/BxN mice, originally described in 1996 as a model of polyarthritis, exhibit knee joint alterations. The aim of this study was to describe temporomandibular joint (TMJ) inflammation and damage in these mice. We used relevant imaging modalities, such as micro-magnetic resonance imaging (μMRI) and micro-computed tomography (μCT), as well as histology and immunofluorescence techniques to detect TMJ alterations in this mouse model. Histology and immunofluorescence for Col-I, Col-II, and aggrecan showed cartilage damage in the TMJ of K/BxN animals, which was also evidenced by μCT but was less pronounced than that seen in the knee joints. μMRI observations suggested an increased volume of the upper articular cavity, an indicator of an inflammatory process. Fibroblast-like synoviocytes (FLSs) isolated from the TMJ of K/BxN mice secreted inflammatory cytokines (IL-6 and IL-1β) and expressed degradative mediators such as matrix metalloproteinases (MMPs). K/BxN mice represent an attractive model for describing and investigating spontaneous damage to the TMJ, a painful disorder in humans with an etiology that is still poorly understood. The volume estimation and 3D reconstructions were obtained with 3D Slicer. |
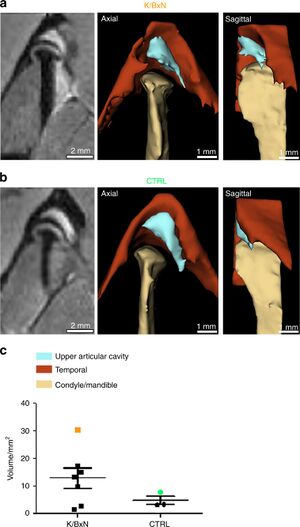 Comparison of synovial fluid volume of K/BxN and control TMJs. Magnetic resonance imaging (MRI) and 3D reconstructions of the TMJ of a K/BxN mouse (a) and a control mouse (b). c Volume measurement of the upper articular cavity of seven K/BxN mice and three control mice. The orange square represents the K/BxN mouse shown in a, and the green circle represents the control mouse shown in b. |
Synchrotron Radiation-Based Reconstruction of the Human Spiral Ganglion: Implications for Cochlear Implantation
|
Publication: Ear Hear. 2020 Jan/Feb;41(1):173-81. PMID: 31008733 Authors: Li H, Schart-Morén N, Rohani SA, Ladak HM, Rask-Andersen H, Agrawal S. Institution: Department of Surgical Sciences, Head and Neck Surgery, Section of Otolaryngology, Uppsala University Hospital, Uppsala, Sweden. Abstract: OBJECTIVE: To three-dimensionally reconstruct Rosenthal's canal (RC) housing the human spiral ganglion (SG) using synchrotron radiation phase-contrast imaging (SR-PCI). Straight cochlear implant electrode arrays were inserted to better comprehend the electro-cochlear interface in cochlear implantation (CI). DESIGN: SR-PCI was used to reconstruct the human cochlea with and without cadaveric CI. Twenty-eight cochleae were volume rendered, of which 12 underwent cadaveric CI with a straight electrode via the round window (RW). Data were input into the 3D Slicer software program and anatomical structures were modeled using a threshold paint tool. RESULTS: The human RC and SG were reproduced three-dimensionally with artefact-free imaging of electrode arrays. The anatomy of the SG and its relationship to the sensory organ (Corti) and soft and bony structures were assessed. CONCLUSIONS: SR-PCI and computer-based three-dimensional reconstructions demonstrated the relationships among implanted electrodes, angular insertion depths, and the SG for the first time in intact, unstained, and nondecalcified specimens. This information can be used to assess stimulation strategies and future electrode designs, as well as create place-frequency maps of the SG for optimal stimulation strategies of the human auditory nerve in CI. |
Visualization of Mucosal Field in HPV Positive and Negative Oropharyngeal Squamous Cell Carcinomas: Combined Genomic and Radiology Based 3D Model
|
Publication: Sci Rep. 2020 Jan 8;10(1):40. PMID: 31913295 | PDF Authors: Orosz E, Gombos K, Petrevszky N, Csonka D, Haber I, Kaszas B, Toth A, Molnar K, Kalacs K, Piski Z, Gerlinger I, Burian A, Bellyei S, Szanyi I. Institution: University of Pécs, Medical School, Department of Otorhinolaryngology, Pécs, Hungary. Abstract: The aim of this study was to visualize the tumor propagation and surrounding mucosal field in radiography-based 3D model for advanced stage HNSCC and combine it with HPV genotyping and miRNA expression characterization of the visualized area. 25 patients with T1-3 clinical stage HNSCC were enrolled in mapping biopsy sampling. Biopsy samples were evaluated for HPV positivity and miR-21-5p, miR-143, miR-155, miR-221-5p expression in Digital Droplet PCR system. Significant miRNA expression differences of HPV positive tumor tissue biopsies were found for miR-21-5p, miR-143 and miR-221-5p compared to the HPV negative tumor biopsy series. Peritumoral mucosa showed patchy pattern alterations of miR-21-5p and miR-155 in HPV positive cases, while gradual change of miR-21-5p and miR-221-5p was seen in HPV negative tumors. In our study we found differences of the miRNA expression patterns among the HPV positive and negative tumorous tissues as well as the surrounding mucosal fields. The CT based 3D models of the cancer field and surrounding mucosal surface can be utilized to improve proper preoperative planning. Complex evaluation of HNSCC tissue organization field can elucidate the clinical and molecular differentiation of HPV positive and negative cases, and enhance effective organ saving therapeutic strategies. |
Using of a Dismountable 3D-model of the Collecting System with Color Segmentation to Improve the Learning Curve of Residents
|
Publication: Urologiia. 2020 Jan;(6):21-5. PMID: 32003162 Authors: Guliev BG, Komyakov BK, Talyshinskiy AE, Stetsik EO. Institution: Department of Urology of FGBOU VO North-Western State Medical University named after I.I. Mechnikov, Saint Petersburg, Russia. Abstract: AIM: to determine the efficiency of using a non-biological dismountable 3D-model of the collecting system with color segmentation for better understanding of its anatomy by residents and to determine the optimal tactics of percutaneous nephrolithotomy (PNL). MATERIALS AND METHODS: 3D-models of the collecting system were developed based on CT data of 5 patients with staghorn stones, for whom PNL was planned. CT images were obtained in the Dicom format. RadiAnt DICOM Viewer was used for delineation and segmentation of the collecting system with 3D visualization. Using 3D Slicer v.4.8.1 software, virtual models were processed to convert DICOM files to STL format. Then, virtual color extraction of each group of calyxes was performed for convenient disassembling and intraluminal study of the anatomy of the collecting system. The final stage included the printing of each area by the method of layer-by-layer deposition using a 3D printer Picaso designer X. To assess the efficiency of the dismountable 3D-model that simulates a certain collecting system, a questionnaire was used. It allowed to evaluate the understanding of the anatomy of the collecting system by residents, as well as the ability to determine the optimal calyx for PNL by comparing the answers with the result of a survey of practicing urologists who had performed more than 50 cases. RESULTS: After studying 3D-models by residents, determination of the number of calyxes in each group was not statistically significantly different from those for practicing urologists who used CT images. The choice of the calyx for primary puncture was not different between groups. However, residents chose the calyx for additional access worse (p=0.009). CONCLUSION: The dismountable 3D-model of the collecting system is promising for training of residents and planning PNL. Studying the anatomy of a single group of calyxes as well as the entire collecting system allows to choose the optimal calyx for percutaneous puncture during PNL. |
Manual and Semiautomatic Segmentation of Bone Sarcomas on MRI Have High Similarity
|
Publication: Braz J Med Biol Res. 2020 Jan 31;53(2):e8962. PMID: 32022102 | PDF Authors: Dionísio FCF, Oliveira LS, Hernandes MA, Engel EE, Rangayyan RM, Azevedo-Marques PM, Nogueira-Barbosa MH. Institution: Departamento de Imagens Médicas, Hematologia e Oncologia Clínica, Faculdade de Medicina de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, SP, Brasil. Abstract: The aims of this study were to evaluate the intra- and interobserver reproducibility of manual segmentation of bone sarcomas in magnetic resonance imaging (MRI) studies and to compare manual and semiautomatic segmentation methods. This retrospective study included twelve osteosarcoma and eight Ewing sarcoma MRI studies performed prior to any therapeutic intervention. All cases were histopathologically confirmed. Three radiologists used 3D Slicer software to perform manual segmentation of bone sarcomas in a blinded and independent manner. One radiologist segmented manually and also performed semiautomatic segmentation with the GrowCut tool. Segmentation exercises were timed for comparison. The dice similarity coefficient (DSC) and Hausdorff distance (HD) were used to evaluate similarity between the segmentation results and further statistical analyses were performed to compare DSC, HD, and volumetric results. Manual segmentation was reproducible with intraobserver DSC varying from 0.83 to 0.97 and HD from 3.37 to 28.73 mm. Interobserver DSC of manual segmentation showed variation from 0.73 to 0.97 and HD from 3.93 to 33.40 mm. Semiautomatic segmentation compared to manual segmentation resulted in DSCs of 0.71-0.96 and HDs of 5.38-31.54 mm. Semiautomatic segmentation required significantly less time compared to manual segmentation (P value ≤0.05). Among all situations compared, tumor volumetry did not show significant statistical differences (P value >0.05). We found excellent intra- and interobserver agreement for manual segmentation of osteosarcoma and Ewing sarcoma. There was high similarity between manual and semiautomatic segmentation, with a significant reduction of segmentation time using the semiautomatic method. |
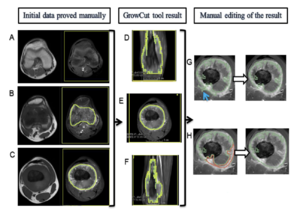 3D Slicer’s GrowCut semiautomatic segmentation steps of a femoral osteosarcoma magnetic resonance imaging case. In the first column (A, B, and C), the T1WI sequence images are on the left and the T1WI FS GD sequence images are on the right. In the second (D, E, and F) and third columns (G and H), there are only T1WI FS GD sequence images. A, Yellow rectangular area of background tissue beyond the inferior margin of tumor. B and C, Area of interest from the tumor tissue within green mark and background tissue within yellow mark in the extremities of the tumor (B) and in the middle portion of the tumor on the longitudinal axis (C). D, E, and F, Segmented volume of interest within green mark after GrowCut tool processing in coronal (D), axial (E), and sagittal (F) planes. G and H, Manual editing of GrowCut tool excluding areas from the volume of interest (within green mark) that do not contain tumor tissue, as indicated with the blue arrow in (G), and including areas with tumor tissue in the volume of interest, as shown in area marked in red (H). |
Measurement Error and Reliability of Three Available 3D Superimposition Methods in Growing Patients
|
Publication: Head Face Med. 2020 Jan 27;16(1):1. PMID: 31987041 | PDF Authors: Ponce-Garcia C, Ruellas ACO, Cevidanes LHS, Flores-Mir C, Carey JP, Lagravere-Vich M. Institution: Division of Orthodontics, School of Dentistry, University of Alberta, Edmonton, Canada. Abstract: Cone-Beam Computed Tomography (CBCT) images can be superimposed, allowing three-dimensional (3D) evaluation of craniofacial growth/treatment effects. Limitations of 3D superimposition techniques are related to imaging quality, software/hardware performance, reference areas chosen, and landmark points/volumes identification errors. The aims of this research are to determine/compare the intra-rater reliability generated by three 3D superimposition methods using CBCT images, and compare the changes observed in treated cases by these methods. METHODS: Thirty-six growing individuals (11-14 years old) were selected from patients that received orthodontic treatment. Before and after treatment (average 24 months apart) CBCTs were analyzed using three superimposition methods. The superimposed scans with the two voxel-based methods were used to construct surface models and quantify differences using 3D SlicerCMF software, while distances in the landmark-derived method were calculated using Excel. 3D linear measurements of the models superimposed with each method were then compared. RESULTS: Repeated measurements with each method separately presented good to excellent intraclass correlation coefficient (ICC ≥ 0.825). ICC values were the lowest when comparing the landmark-based method and both voxel-based methods. Moderate to excellent agreement was observed when comparing the voxel-based methods against each other. The landmark-based method generated the highest measurement error. CONCLUSIONS: Findings indicate good to excellent intra-examiner reliability of the three 3D superimposition methods when assessed individually. However, when assessing reliability among the three methods, ICC demonstrated less powerful agreement. The measurements with two of the three methods (CMFreg/3D Slicer and Dolphin) showed similar mean differences; however, the accuracy of the results could not be determined. |
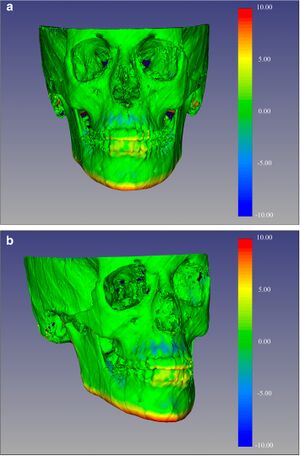 Color-coded map with CMFreg/3D Slicer method for visualization purposes only, not quantitative assessment. Frontal (4a) and 45 degrees (4b) views of the 3D color-coded maps showing the change in mm. |
Prostate Multiparametric Magnetic Resonance Imaging Features Following Partial Gland Cryoablation
|
Publication: Urology. Urology. 2020 Apr;138:98-105. PMID: 31954170 Authors: Al Awamlh BAH, Margolis DJ, Gross MD, Natarajan S, Priester A, Hectors S, Ma X, Mosquera JM, Liao J, Hu JC. Institution: Department of Urology, New York Presbyterian Hospital, Weill Cornell Medicine, New York, NY. Abstract: OBJECTIVES: To assess the qualitative and quantitative changes on prostate multiparametric magnetic resonance imaging (mpMRI) following partial gland ablation (PGA) with cryotherapy and correlate with histopathology. METHODS: We used 3D Slicer to generate prostate models and segment ipsilateral (treated) and contralateral peripheral and transition zones in ten men who underwent MRI/transrectal ultrasound fusion-guided PGA during 2017-2018. Pre and post-PGA volumes of prostate segments were compared. Post-PGA mpMRI were categorized according to PI-RADS v2 and treatment response on mpMRI was assessed in a manner similar to the radiology evaluation framework following liver lesion ablation. RESULTS: Median volume of ipsilateral peripheral and transition zones decreased from 10.9 mL and 13.0 mL to 7.2 mL and 10.8 mL (p=0.005), respectively. Median volume of contralateral peripheral and transition zones also decreased from 12.1 mL and 12.5 mL to 9.9 mL to 10.4 mL (p=0.005), respectively. Five men had clinically significant disease (Grade group ≥2) on post-PGA biopsy (three within treatment field and two outside). Of the men with clinically significant prostate cancer, mpMRI revealed PI-RADS 3 lesions in two. However, the treatment response framework did not detect residual disease. CONCLUSION: PGA results in asymmetric and significant reductions in prostate volume. Our results highlight the need for a separate assessment framework to enable standardization of the interpretation and reporting of post-PGA surveillance mpMRI. Moreover, our findings have significant implications for MRI-targeted surveillance biopsy following PGA with cryotherapy. |
3D Printing Method for Next-Day Acetabular Fracture Surgery Using a Surface Filtering Pipeline: Feasibility and 1-Year Clinical Results
|
Publication: Int J Comput Assist Radiol Surg. 2020 Jan 2. PMID: 31897965 Authors: Weidert S, Andress S, Linhart C, Suero EM, Greiner A, Böcker W, Kammerlander C, Becker CA. Institution: Department of General, Trauma and Reconstructive Surgery, University Hospital, LMU Munich, Campus Großhadern, Munich, Germany. Abstract: INTRODUCTION: In orthopedic surgery, 3D printing is a technology with promising medical applications. Publications show promising results in acetabular fracture surgery over the last years using 3D printing. However, only little information about the workflow and circumstances of how to properly derive the 3D printed fracture model out of a CT scan is published. MATERIALS AND METHODS: We conducted a retrospective analysis of patients with acetabular fractures in a level 1 trauma center. DICOM data were preoperatively used in a series of patients with acetabular fractures. The 3D mesh models were created using 3D Slicer with a newly introduced surface filtering method. The models were printed using PLA material with FDM printer. After reduction in the printed model, the acetabular reconstruction plate was bent preoperatively and sterilized. A clinical follow-up after 12 months in average was conducted with the patients. RESULTS: In total, 12 patients included. Mean printing time was 8:40 h. The calculated mean printing time without applying the surface filter was 25:26 h. This concludes an average printing time reduction of 65%. Mean operation time was 3:16 h, and mean blood loss was 853 ml. Model creation time was about 11 min, and mean printing time of the 3D model was 8:40 h, preoperative model reduction time was 5 min on average, and preoperative bending of the plate took about 10 min. After 12 months, patients underwent a structured follow-up. Harris Hip Score was 75.7 points, the Modified Harris Hip Score 71.6 points and the Merle d'Aubigne Score 11.1 points on average. CONCLUSIONS: We presented the first clinical practical technique to use 3D printing in acetabular fracture surgery. By introducing a new surface filtering pipeline, we reduced printing time and cost compared to the current literature and the state of the art. Low costs and easy handling of the 3D printing workflow make it usable in nearly every hospital setting for acetabular fracture surgery. Funding:
|
Convolutional Neural Network-based MR Image Analysis for Alzheimer's Disease Classification
|
Publication: Curr Med Imaging Rev. 2020;16(1):27-35. PMID: 31989891 Authors: Choi BK, Madusanka N, Choi HK, So JH, Kim CH, Park HG, Bhattacharjee S, Prakash D. Institution: Department of Digital Anti-Aging Healthcare, u-AHRC, Inje University, Gimhae, Korea. Abstract: BACKGROUND: In this study, we used a convolutional neural network (CNN) to classify Alzheimer's disease (AD), mild cognitive impairment (MCI), and normal control (NC) subjects based on images of the hippocampus region extracted from magnetic resonance (MR) images of the brain. METHODS: The datasets used in this study were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI). To segment the hippocampal region automatically, the patient brain MR images were matched to the International Consortium for Brain Mapping template (ICBM) using 3D-Slicer software. Using prior knowledge and anatomical annotation label information, the hippocampal region was automatically extracted from the brain MR images. RESULTS: The area of the hippocampus in each image was preprocessed using local entropy minimization with a bi-cubic spline model (LEMS) by an inhomogeneity intensity correction method. To train the CNN model, we separated the dataset into three groups, namely AD/NC, AD/MCI, and MCI/NC. The prediction model achieved an accuracy of 92.3% for AD/NC, 85.6% for AD/MCI, and 78.1% for MCI/NC. CONCLUSION: The results of this study were compared to those of previous studies, and summarized and analyzed to facilitate more flexible analyses based on additional experiments. The classification accuracy obtained by the proposed method is highly accurate. These findings suggest that this approach is efficient and may be a promising strategy to obtain good AD, MCI and NC classification performance using small patch images of hippocampus instead of whole slide images. |
Go to 2020 :: 2019 :: 2018 :: 2017 :: 2016 :: 2015 :: 2014-2011:: 2010-2005