Difference between revisions of "Main Page"
From Slicer Wiki
| Line 9: | Line 9: | ||
<gallery widths="200px" perrow="4"> | <gallery widths="200px" perrow="4"> | ||
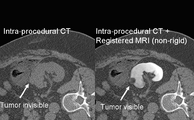
| + | Image:deformable_registration_Screenshot1.png| Non-rigid registration module in 3D Slicer can be used to fuse pre-procedural MR images and intra-procedural CT images during cryoablation of renal tumors. The non-rigid registrations are more accurate than rigid registrations. | ||
Image:LipsMuscles.png|Three-dimensional appearance of the lip muscles with three-dimensional isotropic MRI: An in vivo study. R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. [http://www.slicer.org/slicerWeb/images/4/45/Olszewski-IJCARS2009.pdf See here for the pdf] | Image:LipsMuscles.png|Three-dimensional appearance of the lip muscles with three-dimensional isotropic MRI: An in vivo study. R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. [http://www.slicer.org/slicerWeb/images/4/45/Olszewski-IJCARS2009.pdf See here for the pdf] | ||
Image:Avf 3d voronoi big.png| A Voronoi diagram used for [[Modules:VMTKCenterlines|centerline computation]] of a vascular tree. | Image:Avf 3d voronoi big.png| A Voronoi diagram used for [[Modules:VMTKCenterlines|centerline computation]] of a vascular tree. | ||
Image:LesionsWithVentricles.png|[[Modules:LesionSegmentationApplications-Documentation-3.6|White matter lesion segmentation]] is now available as an extension to Slicer | Image:LesionsWithVentricles.png|[[Modules:LesionSegmentationApplications-Documentation-3.6|White matter lesion segmentation]] is now available as an extension to Slicer | ||
| − | |||
</gallery> | </gallery> | ||
Revision as of 03:24, 3 May 2010
Welcome to the 3D Slicer Wiki pages!
Welcome to the Slicer Wiki Home Page.
How to use this site:
- The main slicer.org pages provide a guided tour to the application, training materials, and the development community. New users should start there because we try to keep it organized and up to date.
- These wiki pages are more free-form, with notes and reference pages that help support developers and users across multiple sites. You will find good information on the wiki, but it's possible that it's out of date or describes features that are still in the planning phase.
Three-dimensional appearance of the lip muscles with three-dimensional isotropic MRI: An in vivo study. R. Olszewski, Y. Liu, T. Duprez, T.M. Xu, H. Reychler: Int J Comput Assist Radiol Surg. 2009 Jun;4(4):349-52. See here for the pdf
A Voronoi diagram used for centerline computation of a vascular tree.
White matter lesion segmentation is now available as an extension to Slicer
More images at the Slicer3 Visual Blog...
In a nutshell, Slicer3 is:
- A cross-platform end user application for analyzing and visualizing medical images.
- A collection of Open Source libraries for developing and deploying new image computing technologies.
- Intended to replace the venerable Slicer2 (see information on the transition)
- An algorithm development platform with a powerful new Execution Model to facilitate creation of new modules.
News
- April 2010
- Slicer factoids on Ohloh
- Feature Freeze month! Developers are working on bug fixes, documentation, and performance enhancement.
- Check out Andras Jakab's impressive surgery planning images
- March 2010
- New infrastructure software is rapidly evolving in the context of the Common Toolkit working meetings.
- Planning underway for release 3.6 of 3D Slicer
- A slicer review on medfloss
- slicer's debian package page
- slicer ubuntu package that can be installed with the alpha version of Ubuntu 10.04
- A page with information about installing slicer3 on Debian has been started.
- We are starting to make plans for 3D Slicer version 4 (also known as Slicer4). A big step!
- January 2010
- December 2009
- Lots of activity around the Port of Slicer to Qt
- New Annotation Tools are in the works
- November 2009
- Seb Barre from Kitware has taken the lead in adding multiple cameras support to Slicer 3. The work is currently focused on multiple cameras and multiple views, but only one scene. The nightly build contains a new module, called cameras. Jim Miller has added a new layout, called Dual 3D Layout. .
- October 2009
- Test of mobile friendly wiki skin
- September 2009
- Serious work is beginning on a Slicer3 port to Qt.
- Attila Nagy and his colleagues created a very nice set of presentations and animations with slicer to illustrate 3D reconstruction as applied to otorhinolaryngology. (See Notes).
- August 2009
- New images in the visual blog show integration of slicer and BioImageSuite.
- July 2009
- Several bug fixes are being rolled into a 3.4.1 patch release, expected for beginning of August
- June 2009
- NA-MIC Summer Project Week was a great success!
- New documentation is available, thanks to the Summer 2009 Tutorial Contest
- May 2009
- Slicer 3.4 release
- The 3.4 release branch has been created in subversion
- A punch list of 3.4 bugs is being maintained on the Mantis Bug Tracker.
- March 2009
- Development of slicer3.4 is still under way. To check the status, go to the bug tracker and search for the tag "3.4 Targeted fix". Our plan is to fix anything with that tag before branching the release. http://na-mic.org/Mantis/main_page.php The svn trunk for slicer3 is still in "feature freeze" mode, meaning that only fixes should be checked in. Any new development should occur in svn branches for now.
- Developers, please be sure to apply the "3.4 Targeted fix" to anything you are working on for the release.
- January 2009
- A product release of Slicer 3.4 is scheduled for Feb/March
- There will be a code freeze on Feb 4, 2009
- Slicer3 and related research project were highlighted and demoed as part of the External Advisory Board meeting of the Neuroimage Analysis Center.
- December 2008
- Andrew Farke posted some excellent tutorials on the use of 3D Slicer in paleontology research.
- A training course on use of 3D Slicer by Radiologists was held at RSNA in Chicago.
- NCIGT is hosting a Project Week for developing Image Guided Therapy software that features many slicer projects.
- September 2008
- Andras Jakab won second place in Kitware's medical image visualization contest!. The image combines slice viewing, models, volume rendering, and tractography applied to a gamma knife neurooncology radiotherapy planning case. Andras used slicer's registration tools to fuse CT and diffusion MR into a multimodal scene.
- A NA-MIC Workshop at MICCAI addressed the creation of ITK-based command line modules for 3D Slicer.
- Slicer user Attila Nagy presented on Slicer3 for inner ear imaging (Slides - Hungarian)
- Attila also provided patches to build Slicer3 and most of Slicer2 using the latest versions of Solaris and the optimized Sun C++ compilers.
- August 2008
- July 2008
- Airway Inspector is a powerful tool for lung image analysis build on slicer2. A port to slicer3 is getting under way in the next few weeks and is expected to take about 6 months.
- Slicer and the NA-MIC software environment were presented as part of the UCLA Institute for Pure and Applied Mathematics Summer School on Mathematics and Brain Imaging.
- A very productive working meeting between researchers at BWH and Yale resulted in successful connection of Slicer3 to the BrainLab commercial image guided neurosurgery system.
- June 2008
- May 2008
- Slicer 3.2 release
- Slicer3 included in review paper on "Rapid Development of Medical Imaging Tools with Open Source Libraries"
- April 2008
- March 2008
- A new 3D Slicer Wikipedia entry has been created.
- Feb 2008
- February 20th, 2008, Spring 2008 Slicer3 User Training Workshop in the SPL at 1249 Boylston St. Boston.
- Jan 2008
- Wishlist for tutorials
- Lots of Slicer development activity as part of NA-MIC All Hands Meeting and Winter Project Week
- December 2007
- A three-day retreat for Slicer in Image Guided Therapy was held in Boston.
- November 2007
- New builds are now available for Linux 64, Linux 32, Windows, Darwin PPC, and Darwin x86
- June 2007
- Lots of Slicer3 work at the Project Weeks
- March 2007
- New Image Guided Therapy (IGT) functionality for tracking surgical instruments added by developers working at the National Center for IGT.
- February 2007
- Feb 7-8, 2007. Slicer3 Mini-Retreat, Boston
- Resulted in the implementation of a Python interface to Slicer3.
- January 2007
- Slicer3 developers are getting together in Salt Lake City for a "programming half-week" as part of the NA-MIC All-Hands Meeting
- Lots of new developer documentation added!
- Slicer3 beta released
- New bug tracker installed
- December 2006
- There was a big meltdown of the na-mic.org server in December, and some things were lost including wiki history, slicer3 bug tracker and doxygen, and some of the older download files. Please bear with us as we pick ourselves up and carry on.
- November 2006
- Slicer3 Alpha builds available for Linux (32 and 64 bit) and windows!
- October 2006
- Slicer3 development version shown at the BIRN All Hands Meeting as the platform for the BIRN Query Atlas Project and overall visualization issue in BIRN. See these slides for more information.
- September 2006:
- July, 2006:
- There was active development of Slicer3 at and around the NA-MIC Programming/Project week.
- New Slicer3 wiki page organization is under way. Please send feedback to the slicer-developers mailing list.
For Wiki Administration
- Create user account
- Users List
- Authorized users can commit edited web pages through slicer's wiki2web interface