Difference between revisions of "EMSegmenter-UseCaseAdvertisement"
From Slicer Wiki
Belhachemi (talk | contribs) |
m (Text replacement - "\[http:\/\/www\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) ([^]]+)]" to "$2") |
||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| + | [[EMSegmenter-Overview|Return to EMSegmenter Overview Page]] <BR> | ||
| + | |||
| + | [[File:EMSegmenterLogo.png|center]] | ||
=Call for Datasets for the EM Segmenter Use Case Library= | =Call for Datasets for the EM Segmenter Use Case Library= | ||
| − | Let us help you turn your segmentation problem into a success story. If you have an interesting | + | Let us help you turn your segmentation problem into a success story. If you have an interesting segmentation problem that is not yet covered in the [[EMSegmenter-Tasks| library ]], send us your case: we will try to segment it for you and design a tailored segmentation approach you can use. We then add the anonymized data to the library and build a guided step-by-step tutorial for it. This is a time and resource limited service available until July 1, 2011. <br> |
If interested drop a line to '''Dominique.Belhachemi at uphs.upenn.edu''' or '''Kilian.Pohl at uphs.upenn.edu''' | If interested drop a line to '''Dominique.Belhachemi at uphs.upenn.edu''' or '''Kilian.Pohl at uphs.upenn.edu''' | ||
| − | |||
| − | |||
=What You Will Need To Do= | =What You Will Need To Do= | ||
* just send us an Email with a brief description of the problem | * just send us an Email with a brief description of the problem | ||
| − | * send us your image data with the corresponding manual segmentation. Please provide more than 10 instances, so that we can generate a robust atlas. | + | * send us your image data with the corresponding (manual) segmentation. Please provide more than 10 instances, so that we can generate a robust atlas. Please also insure that the data can be publicly distributed |
* save your image data as NRRD or NIFTI files | * save your image data as NRRD or NIFTI files | ||
* if you are unsuccessful in saving the data as described above (which is the anonymization), see our [http://wiki.na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:Anonymization Anonymization Instructions] , or let us know and we'll walk you through it. | * if you are unsuccessful in saving the data as described above (which is the anonymization), see our [http://wiki.na-mic.org/Wiki/index.php/Projects:RegistrationDocumentation:Anonymization Anonymization Instructions] , or let us know and we'll walk you through it. | ||
| Line 15: | Line 16: | ||
=What We Will Do= | =What We Will Do= | ||
| − | * based on your | + | * based on your segmentations we will create an atlas |
* we will create a 3DSlicer scene with optimized EM Segmenter parameters for your use case | * we will create a 3DSlicer scene with optimized EM Segmenter parameters for your use case | ||
* we will send you the final result as well as the procedure and parameters | * we will send you the final result as well as the procedure and parameters | ||
| − | * we will add your use case to our | + | * we will add your use case (3DSlicer scene + atlas data) to our [[EMSegmenter-Tasks|Segmentation Task Library]] which will become part of a future 3DSlicer version |
* this will contain parameters for a successful segmentation which you can load directly into slicer and apply on your data | * this will contain parameters for a successful segmentation which you can load directly into slicer and apply on your data | ||
| − | + | * this page will also contain a description of your segmentation problem, the exact workflow of obtaining the segmentation, an acknowledgment of your lab and links to your institution and related research paper | |
| − | * this page will contain a description of your segmentation problem, the exact workflow of obtaining the segmentation, an acknowledgment of your lab and links to your institution and related research paper | ||
* if you can provide us with criteria that define a good segmentation, we will use them in optimization efforts. | * if you can provide us with criteria that define a good segmentation, we will use them in optimization efforts. | ||
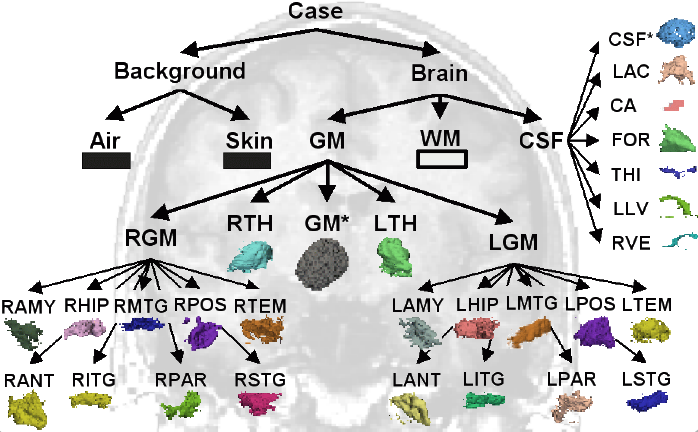
| − | * example of what the final shared product will look like | + | * [[ EMSegmenter-Tasks:MRI-Human-Brain-Parcellation | example of what the final shared product will look like ]] |
| − | |||
=Limitations/Disclaimer= | =Limitations/Disclaimer= | ||
| Line 31: | Line 30: | ||
* this is a time-limited service, available in a first trial until 'July 1, 2011. | * this is a time-limited service, available in a first trial until 'July 1, 2011. | ||
* depending on workload, we try to get you something within a few weeks, but we cannot guarantee a particular result or a specific turnaround time. | * depending on workload, we try to get you something within a few weeks, but we cannot guarantee a particular result or a specific turnaround time. | ||
| − | * all shared data-sets are anonymized | + | * all shared data-sets are anonymized. Sharing anonymized data in most cases is IRB exempt based on Code of Federal Regulations 45 CFR 46.101(b), which lists as exemption category 4: |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | Unless otherwise required by department or agency heads, [...] the following categories are exempt [...]: | |
| − | + | ||
| − | + | 4.Research involving the collection or study of existing data, documents, records , pathological specimens, | |
| + | or diagnostic specimens, if these sources are publicly available or if the information is recorded by the | ||
| + | investigator in such a manner that subjects cannot be identified, directly or through identifiers linked | ||
| + | to the subjects. | ||
Latest revision as of 02:25, 27 November 2019
Home < EMSegmenter-UseCaseAdvertisementReturn to EMSegmenter Overview Page
Contents
Call for Datasets for the EM Segmenter Use Case Library
Let us help you turn your segmentation problem into a success story. If you have an interesting segmentation problem that is not yet covered in the library , send us your case: we will try to segment it for you and design a tailored segmentation approach you can use. We then add the anonymized data to the library and build a guided step-by-step tutorial for it. This is a time and resource limited service available until July 1, 2011.
If interested drop a line to Dominique.Belhachemi at uphs.upenn.edu or Kilian.Pohl at uphs.upenn.edu
What You Will Need To Do
- just send us an Email with a brief description of the problem
- send us your image data with the corresponding (manual) segmentation. Please provide more than 10 instances, so that we can generate a robust atlas. Please also insure that the data can be publicly distributed
- save your image data as NRRD or NIFTI files
- if you are unsuccessful in saving the data as described above (which is the anonymization), see our Anonymization Instructions , or let us know and we'll walk you through it.
What We Will Do
- based on your segmentations we will create an atlas
- we will create a 3DSlicer scene with optimized EM Segmenter parameters for your use case
- we will send you the final result as well as the procedure and parameters
- we will add your use case (3DSlicer scene + atlas data) to our Segmentation Task Library which will become part of a future 3DSlicer version
- this will contain parameters for a successful segmentation which you can load directly into slicer and apply on your data
- this page will also contain a description of your segmentation problem, the exact workflow of obtaining the segmentation, an acknowledgment of your lab and links to your institution and related research paper
- if you can provide us with criteria that define a good segmentation, we will use them in optimization efforts.
- example of what the final shared product will look like
Limitations/Disclaimer
- we can assist only with 3DSlicer software.
- this is a time-limited service, available in a first trial until 'July 1, 2011.
- depending on workload, we try to get you something within a few weeks, but we cannot guarantee a particular result or a specific turnaround time.
- all shared data-sets are anonymized. Sharing anonymized data in most cases is IRB exempt based on Code of Federal Regulations 45 CFR 46.101(b), which lists as exemption category 4:
Unless otherwise required by department or agency heads, [...] the following categories are exempt [...]: 4.Research involving the collection or study of existing data, documents, records , pathological specimens, or diagnostic specimens, if these sources are publicly available or if the information is recorded by the investigator in such a manner that subjects cannot be identified, directly or through identifiers linked to the subjects.