Difference between revisions of "EMSegmenter-Tasks:Non-Human-Primate"
| (14 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
=Description= | =Description= | ||
| − | Non Human Primate pipeline ... | + | Non Human Primate pipeline facilitates segmentation of single-channel (T1w) MRI of the vervet brain. The current segmentation pipeline task operates on the skull-stripped brain MRI. In our experience, skull-stripping of NHP brain can be performed by registration of a subject with manually defined ICC to the subject being analyzed. We recommend using [[Modules:BRAINSFit|BRAINSFit]] module for registering ICC template to the subject. In order to improve the robustness of the registration process we recommend prior bias correction using [[Modules:N4ITKBiasFieldCorrection-Documentation-3.6|N4ITKBiasFieldCorrection]] module. The resulting ICC may need to be manually adjusted using [[Modules:Editor-Documentation-3.6|Editor]] module. In the future, we plan to substitute this registration-based skull-stripping approach with the [http://www.na-mic.org/Wiki/index.php/2010_Winter_Project_Week_SPECTRE_3DSlicer_Integration SPECTRE Slicer plugin] currently under development. |
| + | The pipeline is using unbiased atlas of vervet brain. The atlas construction process was facilitated by the unbiased template construction approach implemented in [http://www.picsl.upenn.edu/ANTS/ ANTS] software. The atlas construction is described in the paper [in preparation]. The T1w MR images used in the atlas construction were acquired from the population of 10 male vervets ~4 y.o. The high-level atlas construction approach is the following: | ||
| + | # scan of one subject (S1) was selected | ||
| + | # an estimate of GM/WM/CSF segmentation was obtained using k-means clustering, followed by manual editing as necessary | ||
| + | # subcortical structures (Caudate, Hippocampus and Putamen) were segmented manually | ||
| + | # average template was obtained using [http://www.picsl.upenn.edu/ANTS/templatetut/tutorial.php ANTS Template building approach] | ||
| + | # the inverses of the transforms produced in the previous step for each subjects were applied to the manual segmentation obtained in Step 2 | ||
| + | # the segmentations obtained in the previous step were pixel-wise averaged for each structure to obtain probabilistic atlases | ||
| − | The pipeline consist of the following steps: | + | The current segmentation pipeline consist of the following steps: |
| − | * Step 1 | + | * Step 1: Register the atlas to the MRI scan via [[Modules:BRAINSFit| BRAINSFit]] (Johnson et al 2007) |
| − | + | * Step 2: Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas. | |
| − | * Step | + | * Step 3: Automatically segment the MRI scan into the structures of interest using [[Modules:EMSegmenter-3.6|EM Algorithm]] (Pohl et al 2007) |
| − | Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas. | ||
| − | * Step | ||
=Anatomical Tree= | =Anatomical Tree= | ||
| Line 39: | Line 44: | ||
Image Spacing = 0.4688 x 0.4688 x 0.50002 | Image Spacing = 0.4688 x 0.4688 x 0.50002 | ||
| + | Images were acquired at Wake Forest University. Acquisition details for the images used in the atlas construction: Axial T1-weighted MR images were acquired on a 3T GE scanner with a circularly-polarized, single channel dedicated RF coil with an internal diameter of 18.4 cm (Litzcage, Doty Scientific, Columbia, SC), using a 3D SPGR sequence (TI 600ms, TE 3.276ms, TR 15.28ms; flip angle 15o; matrix 256x256; FOV 12cm; in-plane resolution 0.47mm; slice thickness 0.5mm). | ||
{| class="wikitable" | {| class="wikitable" | ||
| Line 54: | Line 60: | ||
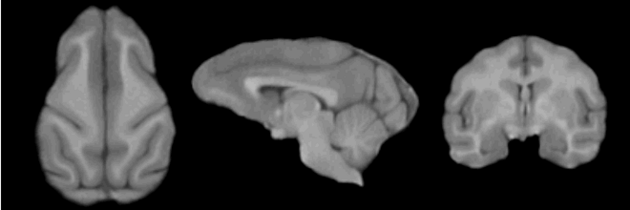
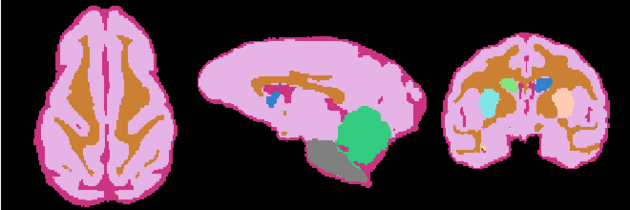
=Result= | =Result= | ||
| − | + | <gallery perrow=1: widths=630px : heights=210px> | |
| − | + | Image:EMSegmenter NHP T1.png | |
| + | Image:NHP_Labelmap.png | ||
| + | </gallery> | ||
| + | |||
| + | =Acknowledgment= | ||
| + | The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement and by R01 AA016748 [http://wiki.na-mic.org/Wiki/index.php/NA-MIC_NCBC_Collaboration:Measuring_Alcohol_and_Stress_Interaction (NA-MIC NCBC Collaboration with Wake Forest University and Virginia Tech: Measuring Alcohol and Stress Interaction)]. | ||
=Collaborators= | =Collaborators= | ||
| − | Andriy Fedorov (BWH) | + | * James Daunais (WFU) |
| + | * Chris Wyatt (VT) | ||
| + | * Bob Croft (WFU) | ||
| + | * Andriy Fedorov (BWH) | ||
| + | * Ron Kikinis (BWH) | ||
=Citations= | =Citations= | ||
| Line 65: | Line 80: | ||
* S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001. | * S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001. | ||
* Johnson H.J., Harris G., Williams K. [http://hdl.handle.net/1926/1291 BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit], The Insight Journal, July 2007 | * Johnson H.J., Harris G., Williams K. [http://hdl.handle.net/1926/1291 BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit], The Insight Journal, July 2007 | ||
| + | * A.Fedorov, X.Li, K.Pohl, S.Bouix, M.Styner, M.Addicott, C.Wyatt, J.B.Daunais, W.M.Wells, R.Kikinis "Atlas-guided Segmentation of Vervet Monkey Brain MRI". The Open Neuroimaging Journal, 2011, 5(Suppl 2-M7), pp.186-197. doi:10.2174/1874440001105010186 [http://www.na-mic.org/publications/item/view/2119 URL] | ||
| + | * NITRC Vervet Brain Atlas [http://www.nitrc.org/projects/vervet_atlas URL] | ||
Latest revision as of 15:48, 7 December 2011
Home < EMSegmenter-Tasks:Non-Human-PrimateReturn to EMSegmenter Task Overview Page
Contents
Description
Non Human Primate pipeline facilitates segmentation of single-channel (T1w) MRI of the vervet brain. The current segmentation pipeline task operates on the skull-stripped brain MRI. In our experience, skull-stripping of NHP brain can be performed by registration of a subject with manually defined ICC to the subject being analyzed. We recommend using BRAINSFit module for registering ICC template to the subject. In order to improve the robustness of the registration process we recommend prior bias correction using N4ITKBiasFieldCorrection module. The resulting ICC may need to be manually adjusted using Editor module. In the future, we plan to substitute this registration-based skull-stripping approach with the SPECTRE Slicer plugin currently under development.
The pipeline is using unbiased atlas of vervet brain. The atlas construction process was facilitated by the unbiased template construction approach implemented in ANTS software. The atlas construction is described in the paper [in preparation]. The T1w MR images used in the atlas construction were acquired from the population of 10 male vervets ~4 y.o. The high-level atlas construction approach is the following:
- scan of one subject (S1) was selected
- an estimate of GM/WM/CSF segmentation was obtained using k-means clustering, followed by manual editing as necessary
- subcortical structures (Caudate, Hippocampus and Putamen) were segmented manually
- average template was obtained using ANTS Template building approach
- the inverses of the transforms produced in the previous step for each subjects were applied to the manual segmentation obtained in Step 2
- the segmentations obtained in the previous step were pixel-wise averaged for each structure to obtain probabilistic atlases
The current segmentation pipeline consist of the following steps:
- Step 1: Register the atlas to the MRI scan via BRAINSFit (Johnson et al 2007)
- Step 2: Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas.
- Step 3: Automatically segment the MRI scan into the structures of interest using EM Algorithm (Pohl et al 2007)
Anatomical Tree
- Root
- Head
- intracranial cavity (ICC)
- White matter + Gray matter + CSF class (WGC)
- grey matter (GM)
- left Subcortical Gray Matter (lSGM)
- left caudate (lCaudate)
- left putamen (lPutamen)
- left hippocampus (lHippocampus)
- right Subcortical Gray Matter (rSGM)
- right caudate (rCaudate)
- right putamen (rPutamen)
- right hippocampus (rHippocampus)
- Cortical Gray Matter (CGM)
- left Subcortical Gray Matter (lSGM)
- white matter (WM)
- cerebrospinal fluid (CSF)
- grey matter (GM)
- Brainstem
- Cerebellum
- White matter + Gray matter + CSF class (WGC)
- background (BG)
- intracranial cavity (ICC)
- Head
Atlas
Image Dimension = 256 x 256 x 252
Image Spacing = 0.4688 x 0.4688 x 0.50002
Images were acquired at Wake Forest University. Acquisition details for the images used in the atlas construction: Axial T1-weighted MR images were acquired on a 3T GE scanner with a circularly-polarized, single channel dedicated RF coil with an internal diameter of 18.4 cm (Litzcage, Doty Scientific, Columbia, SC), using a 3D SPGR sequence (TI 600ms, TE 3.276ms, TR 15.28ms; flip angle 15o; matrix 256x256; FOV 12cm; in-plane resolution 0.47mm; slice thickness 0.5mm).
|
|
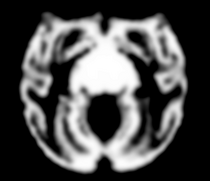
|

|
| Template (T1) | CSF | GM | WM |
Result
Acknowledgment
The construction of the pipeline was supported by funding from NIH NCRR 2P41RR013218 Supplement and by R01 AA016748 (NA-MIC NCBC Collaboration with Wake Forest University and Virginia Tech: Measuring Alcohol and Stress Interaction).
Collaborators
- James Daunais (WFU)
- Chris Wyatt (VT)
- Bob Croft (WFU)
- Andriy Fedorov (BWH)
- Ron Kikinis (BWH)
Citations
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.
- S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001.
- Johnson H.J., Harris G., Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, The Insight Journal, July 2007
- A.Fedorov, X.Li, K.Pohl, S.Bouix, M.Styner, M.Addicott, C.Wyatt, J.B.Daunais, W.M.Wells, R.Kikinis "Atlas-guided Segmentation of Vervet Monkey Brain MRI". The Open Neuroimaging Journal, 2011, 5(Suppl 2-M7), pp.186-197. doi:10.2174/1874440001105010186 URL
- NITRC Vervet Brain Atlas URL