Difference between revisions of "EMSegmenter-Tasks:MRI-Human-Brain-Parcellation"
Belhachemi (talk | contribs) |
Belhachemi (talk | contribs) |
||
| Line 1: | Line 1: | ||
=Description= | =Description= | ||
| − | MRI Human Head pipeline for a finer-grained parcellation (Priya please provide description and proper citation) | + | MRI Human Head pipeline for a finer-grained parcellation ('''TODO''': Priya please provide description and proper citation) |
| + | |||
| + | The pipeline consist of the following steps: | ||
| + | * Step 1: Perform image inhomogeneity correction of the MRI scan via [http://www.slicer.org/slicerWiki/index.php/Modules:N4ITKBiasFieldCorrection-Documentation-3.6 N4ITKBiasFieldCorrection] (Tustison et al 2010) | ||
| + | * Step 2: Register the atlas to the MRI scan via [[Modules:BRAINSFit| BRAINSFit]] (Johnson et al 2007) | ||
| + | * Step 3: Compute the intensity distributions for each structure <BR> | ||
| + | Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas. | ||
| + | * Step 4: Automatically segment the MRI scan into the structures of interest using [[Modules:EMSegmenter-3.6|EM Algorithm]] (Pohl et al 2007) | ||
=Anatomical Tree= | =Anatomical Tree= | ||
| Line 42: | Line 49: | ||
'''TODO, some details about atlas''' | '''TODO, some details about atlas''' | ||
| − | + | =Result= | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
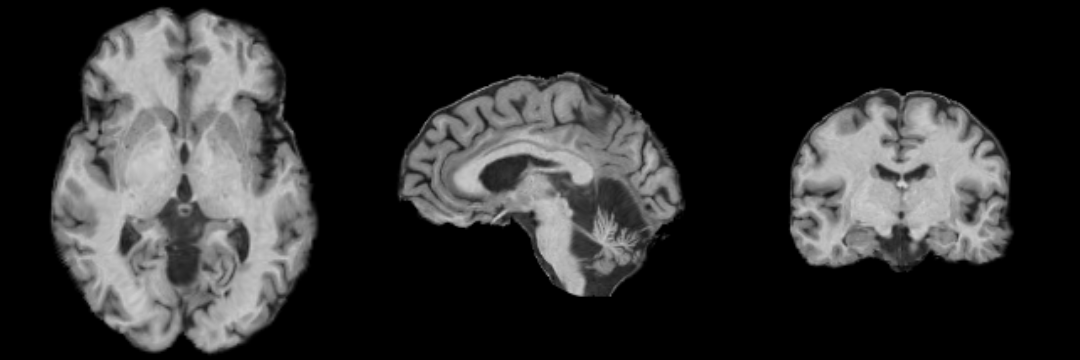
[[Image:MRI-Human-Brain-Parcellation_T1_3x360x360.png]] | [[Image:MRI-Human-Brain-Parcellation_T1_3x360x360.png]] | ||
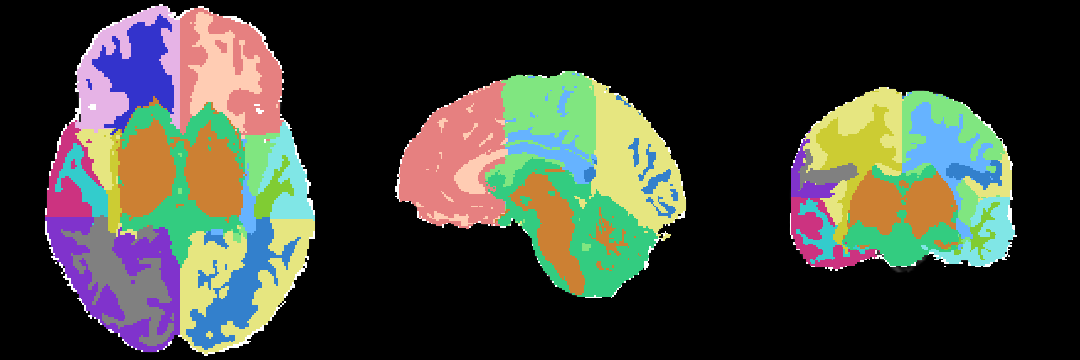
[[Image:MRI-Human-Brain-Parcellation_3x360x360.png]] | [[Image:MRI-Human-Brain-Parcellation_3x360x360.png]] | ||
Revision as of 21:52, 1 December 2010
Home < EMSegmenter-Tasks:MRI-Human-Brain-ParcellationDescription
MRI Human Head pipeline for a finer-grained parcellation (TODO: Priya please provide description and proper citation)
The pipeline consist of the following steps:
- Step 1: Perform image inhomogeneity correction of the MRI scan via N4ITKBiasFieldCorrection (Tustison et al 2010)
- Step 2: Register the atlas to the MRI scan via BRAINSFit (Johnson et al 2007)
- Step 3: Compute the intensity distributions for each structure
Compute intensity distribution (mean and variance) for each label by automatically sampling from the MR scan. The sampling for a specific label is constrained to the region that consists of voxels with high probability (top 95%) of being assigned to the label according to the aligned atlas.
- Step 4: Automatically segment the MRI scan into the structures of interest using EM Algorithm (Pohl et al 2007)
Anatomical Tree
The anatomical tree represents the structures to be segmented. Node labels displayed below contain a human readable structure name and in parentheses the internally used structure name.
- Root
- background (BG)
- grey matter (GM)
- left temporal grey matter (LTGM)
- left temporal grey matter - region 1 (LTGM1)
- left temporal grey matter - region 2 (LTGM2)
- left temporal grey matter - region 3 (LTGM3)
- left temporal grey matter - region 4 (LTGM4)
- right temporal grey matter (RTGM)
- right temporal grey matter - region 1 (RTGM1)
- right temporal grey matter - region 2 (RTGM2)
- right temporal grey matter - region 3 (RTGM3)
- right temporal grey matter - region 4 (RTGM4)
- subcortical grey matter (SUBGM)
- left temporal grey matter (LTGM)
- white matter (WM)
- left temporal white matter (LTWM)
- left temporal white matter - region 1 (LTWM1)
- left temporal white matter - region 2 (LTWM2)
- left temporal white matter - region 3 (LTWM3)
- left temporal white matter - region 4 (LTWM4)
- right temporal white matter (RTWM)
- right temporal white matter - region 1 (RTWM1)
- right temporal white matter - region 2 (RTWM2)
- right temporal white matter - region 3 (RTWM3)
- right temporal white matter - region 4 (RTWM4)
- subcortical white matter (SUBWM)
- left temporal white matter (LTWM)
- cerebrospinal fluid (CSF)
Atlas
The atlas hes been created by Padmapriya Srinivasan and Sylvain Bouix from (PNL-BWH)
Image Dimension = 256 x 256 x 220
Image Spacing = 0.9375 x 0.9375 x 0.9375
TODO, some details about atlas
Result
Collaborators
Padmapriya Srinivasan and Sylvain Bouix (PNL-BWH)
Citations
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC N4ITK: Improved N3 Bias Correction, IEEE Trans Med Imag, 2010
- Pohl K, Bouix S, Nakamura M, Rohlfing T, McCarley R, Kikinis R, Grimson W, Shenton M, Wells W. A Hierarchical Algorithm for MR Brain Image Parcellation. IEEE Transactions on Medical Imaging. 2007 Sept;26(9):1201-1212.
- S. Warfield, J. Rexilius, P. Huppi, T. Inder, E. Miller, W. Wells, G. Zientara, F. Jolesz, and R. Kikinis, “A binary entropy measure to assess nonrigid registration algorithms,” in MICCAI, LNCS, pp. 266–274, Springer, October 2001.
- Johnson H.J., Harris G., Williams K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit, The Insight Journal, July 2007