Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C50"
(Created page with 'Back to Registration Library <br> = Slicer Registration Library Case #50: Mouse micro-CT = == Input == {| style="color…') |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 20: | Line 20: | ||
== Download (from NAMIC MIDAS) == | == Download (from NAMIC MIDAS) == | ||
<small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | ||
| − | *[http://slicer.kitware.com/midas3/download/?items= | + | *[http://slicer.kitware.com/midas3/download/?items=127956 '''RegLib_C50.mrb''': input data only, use this to run the tutorial from the start <small>(Slicer mrb file. 50 MB). </small>] |
| − | *[http://slicer.kitware.com/midas3/download/?items= | + | *[http://slicer.kitware.com/midas3/download/?items=127955 '''RegLib_C50_full.mrb''': includes raw data + all solutions and intermediate files, use to browse/verify <small>(Slicer mrb file. 82 MB). </small>] |
== Keywords == | == Keywords == | ||
| Line 57: | Line 57: | ||
{|cellpadding="10" cellspacing="0" border="0" | {|cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image:RegLib_C50_unregistered.png|300px]] | + | |[[Image:RegLib_C50_unregistered.png|300px]] before registration (click to enlarge) |
|- | |- | ||
| − | |[[Image: | + | |[[Image:RegLib_C50_registered_BSpline.gif|300px]] after affine+nonrigid alignment (click to enlarge) |
|} | |} | ||
=== Acknowledgments === | === Acknowledgments === | ||
| − | Many thanks to Dr.Murat Maga | + | Many thanks to Dr.Murat Maga from [http://www.seattlechildrens.org/research/developmental-biology-regenerative-medicine Center for Developmental Biology and Regenerative Medicine in Seattle, WA] for sharing the example data. |
Latest revision as of 20:30, 23 January 2014
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C50Contents
Slicer Registration Library Case #50: Mouse micro-CT
Input
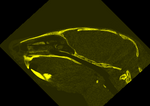
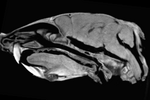
| fixed image/target baseline CT |
moving image CT after fixation + staining |
Description
Goal is to align the two CT images pre/post fixation. The fixation process altered the tissue appearance and also caused distortions we seek to correct with non-rigid registration.
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C50.mrb: input data only, use this to run the tutorial from the start (Slicer mrb file. 50 MB).
- RegLib_C50_full.mrb: includes raw data + all solutions and intermediate files, use to browse/verify (Slicer mrb file. 82 MB).
Keywords
micro-CT, animal, mouse, brain, intra-subject,
Video Screencasts
- RegLib_C50 Registration Screencast (I. Affine Registration)
- RegLib_C50 Registration Screencast (II. BSpline Registration)
Procedure
- Affine Register
- go to the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: baseline
- Input Images: Moving Image Volume: stained
- Output Settings:
- Slicer Linear Transform (create new transform, rename to: "Xf1_Affine")
- Output Image Volume none. No resampling required for linear transforms
- Registration Phases: select/check Rigid , Rigid+Scale, Affine
- Main Parameters:
- increase Number Of Samples to 300,000
- click: Apply; runtime ~ 1 min.
- BSpline Register
- leave baseline and stained as fixed and moving volumes
- Output Settings:
- Slicer Linear Transform select None
- Slicer BSpline Transform create new, rename to "Xf2_BSpline" or similar
- Output Image Volume: create new and rename to "stained_Xf2". Resampling is required for nonrigid transforms.
- Registration Phases: unchheck Rigid , Rigid+Scale, Affine and check "BSpline" only
- Main Parameters:
- increase Number Of Samples to at least 300,000
- BSpline Grid Size: set to 7,7,7 or similar.
- click: Apply; runtime < 3 min.
Registration Results
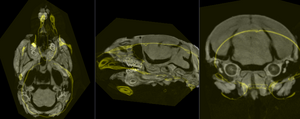 before registration (click to enlarge) before registration (click to enlarge)
|
 after affine+nonrigid alignment (click to enlarge) after affine+nonrigid alignment (click to enlarge)
|
Acknowledgments
Many thanks to Dr.Murat Maga from Center for Developmental Biology and Regenerative Medicine in Seattle, WA for sharing the example data.