Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C44"
(Created page with ' = Slicer Registration Library Case 44: Visible Human Pelvis CT = == Input == {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" |[[Image:RegLib_C44_Thumb1.…') |
(→Input) |
||
| (25 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Documentation/Nightly/Registration/RegistrationLibrary|Back to Registration Library]] | |
= Slicer Registration Library Case 44: Visible Human Pelvis CT = | = Slicer Registration Library Case 44: Visible Human Pelvis CT = | ||
== Input == | == Input == | ||
{| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | ||
|[[Image:RegLib_C44_Thumb1.png|150px|lleft|this is the main fixed reference image. All images are ev. aligned into this space]] | |[[Image:RegLib_C44_Thumb1.png|150px|lleft|this is the main fixed reference image. All images are ev. aligned into this space]] | ||
| − | |[[Image: | + | |[[Image:RegArrow_NonRigid.png|70px|lleft]] |
|[[Image:RegLib_C44_Thumb2.png|150px|lleft|this is the moving image]] | |[[Image:RegLib_C44_Thumb2.png|150px|lleft|this is the moving image]] | ||
|- | |- | ||
| Line 12: | Line 12: | ||
|} | |} | ||
| − | == Modules | + | == Modules used == |
| − | *''' | + | *[[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']] |
| + | *[[Documentation/Nightly/Modules/DeformationFieldVisualizer| ''Deformation Field Visualizer'']] | ||
| − | + | == Description == | |
| − | This dataset contains CT of the visible human male and female pelvis. This serves as a test example for exploring non-rigid registration for inter-subject comparison from CT. | + | This dataset contains CT of the visible human male and female pelvis. This serves as a test example for exploring non-rigid registration for inter-subject comparison from CT. The overall strategy will be to register "vhf" to "vhm" via first affine and then BSpline registration. We will generate a mask to focus the registration on the bone structure only and ignore the soft tissue when computing the deformation. Because our original images are quite large (512x512x150), we will subsample the vhf pair for use with the [[Documentation/Nightly/Modules/DeformationFieldVisualizer| ''Deformation Field Visualizer'']] module, which might otherwise become too memory intensive. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Download == | ==Download == | ||
| − | + | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | |
| − | * | + | *[http://slicer.kitware.com/midas3/download/?items=119555 '''RegLib_C44.mrb'''] <small>(input data only, Slicer mrb file. 74 MB) </small><br> |
| − | + | *[http://slicer.kitware.com/midas3/download/?items=119554 '''RegLib_C44_full.mrb''']<small>(input data + results, Slicer mrb file. 82 MB). </small> | |
| − | + | ||
| + | == Video Screencasts == | ||
| + | #[[Media:RegLib_C44_1_Masking.mov|Movie/screencast showing generating a registration mask]] | ||
| + | #[[Media:RegLib_C44_2_Registration.mov|Movie/screencast showing affine and nonrigid BSpline registration]] | ||
| + | #[[Media:RegLib_C44_3_Deformation.mov|Movie/screencast showing visualization of the deformation via the Transform Visualizer module]] | ||
== Keywords == | == Keywords == | ||
| Line 37: | Line 33: | ||
== Procedure / Pipeline == | == Procedure / Pipeline == | ||
| − | #Mask generation | + | #'''Mask generation''': open the [[Documentation/Nightly/Modules/Editor|''Editor'']] module. [[Media:RegLib_C44_1_Masking.mov|Movie/screencast showing this step]] |
| − | |||
##"Master Volume": select ''vhm'' | ##"Master Volume": select ''vhm'' | ||
##A new labelmap "vhm-label" will be created | ##A new labelmap "vhm-label" will be created | ||
##Select "vhm" to be visible in the slice viewer | ##Select "vhm" to be visible in the slice viewer | ||
##Select the ''Threshold'' tool from the editor toolbar | ##Select the ''Threshold'' tool from the editor toolbar | ||
| − | ##Adjust the lower threshold (slider bar) until most of the bone is highlighted, e.g. somewhere around an intensity value of 80. Leave the upper threshold unchanged at the max. | + | ##Adjust the lower threshold (slider bar) until most of the bone is highlighted,just before speckle noise starts to become included e.g. somewhere around an intensity value of 80. Leave the upper threshold unchanged at the max. |
##Click Apply | ##Click Apply | ||
| − | ## | + | ##clean the segmentation: |
| − | ## | + | ##Select the "Identify Islands" editor effect. [[Image:IdentifyIslandsEffectIcon.png|30px]] This will identify all continuous areas that are disconnected from each other. Click "Apply" |
| − | ## | + | ##you should see the bones of the arms being assigned a different label value & color. We can now delete them with one click: |
| − | ## | + | ##select the "Change Island" effect [[Image:ChangeIslandsEffectIcon.png|30px]] . Change the label value to 0 (zero). |
| − | ## | + | ##in the axial (red) view, click the left mouse within the segmented areas of the arms. |
| − | ## | + | ##select the "Dilate" effect [[Image:DilateEffectIcon.png|30px]]. Click the "Apply" button 3-4 times until the boundary of the segmentation extends well beyond the bone, including a several pixel wide layer of adjacent tissue. |
| − | # | + | ##repeat the above for "vhf". |
| − | + | ##save the two segmentations. | |
| − | + | #'''Affine Registration''': open the [[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']] module. [[Media:RegLib_C44_2_Registration.mov|Movie/screencast showing this step]] | |
| − | + | ##''Fixed Image Volume'': vhm | |
| − | + | ##''Moving Volume'': vhf | |
| − | + | ##check boxes for ''Include Rigd registr. phase'' , ''Include ScaleVersor3D'', ''include Affine'' | |
| − | + | ##''Slicer Linear Transform'': select "create new transform", rename to "Xf1_Affine" or similar | |
| − | + | ##leave rest at defaults. Click ''Apply'' | |
| − | + | ##registration should take ~ 10 secs. | |
| − | + | ##use fade slider to verify alignment; compare with result snapshots shown below. Alignment will not be perfect but should be better than before. | |
| − | + | ##note: you can also change the colormaps for the fixed and moving volumes to better judge the alignment: go to the [[Documentation/Nightly/Modules/Volumes| ''Volumes'']] module and in the ''Display'' tab, select "green" and "magenta" as the respective colormaps for the two volumes (vhf, vhm) | |
| − | # | + | #'''Nonrigid Registration (masked)''': open the [[Documentation/Nightly/Modules/BRAINSFit| ''General Registration (BRAINS)'']]. [[Media:RegLib_C44_2_Registration.mov|Movie/screencast showing this step]] |
| − | # | + | ##''Fixed Image Volume'': vhm ''Moving'': vhf |
| − | + | ##Registration phases: from ''Initialize with previously generated transform', select "Xf1_Affine" node created before. | |
| − | + | ##Registration phases: uncheck boxes for rigid, scale and affine and check box for ''BSpline'' | |
| − | ##Registration phases: from ''Initialize with previously generated transform', select " | + | ##Output: Slicer Linear transform: set to None |
| − | ##Registration phases: check | ||
##Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline_msk" or similar | ##Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline_msk" or similar | ||
##Output Image Volume: create new, rename to "vhf_Xf2"; ''Pixel Type'': "short" | ##Output Image Volume: create new, rename to "vhf_Xf2"; ''Pixel Type'': "short" | ||
| Line 74: | Line 68: | ||
##Leave all other settings at default | ##Leave all other settings at default | ||
##click apply | ##click apply | ||
| + | #'''Deformation Visualization''': [[Media:RegLib_C44_3_Deformation.mov|Movie/screencast showing this step]] | ||
| + | ##if you have not yet installed the [[Documentation/Nightly/Modules/DeformationFieldVisualizer| ''Deformation Field Visualizer'']] extension, [[Documentation/Nightly/RegistrationVideoTutorials#Registration_Validation__How_do_I_check_.2F_visualize_a_registration_result_.3F|see here]] for a movie clip on how to install it. | ||
| + | ##we first generate a smaller version of our image to save memory: | ||
| + | ###open the '''Resize Image (BRAINS)''' module (under Registration) | ||
| + | ###''Image To Warp'': select "vhf" | ||
| + | ###''Output Image'': create new, rename to "vhf_small" or similar | ||
| + | ###''Pixel Type'': select "short" | ||
| + | ###''Scale Factor'': leave at 2.0 | ||
| + | ###click ''Apply'' | ||
| + | ##open the [[Documentation/Nightly/Modules/DeformationFieldVisualizer| ''Transform Visualizer'']] module (under: All Modules) | ||
| + | ###Deformation: select "Xf2_BSpline" | ||
| + | ###Reference Image: select "vhf_small" generated above | ||
| + | ###Visualization mode: click on "Grid Slice" | ||
| + | ###Grid Slice Options: Slice: red; Spacing: 20mm | ||
| + | ###Click Apply. you should see the deformation field overlay. Adjust slice, spacing etc. to taste. | ||
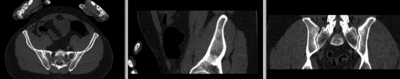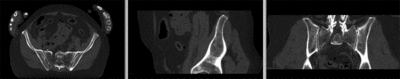
== Registration Results== | == Registration Results== | ||
[[Image:RegLib_C44_unregistered.gif|400px|original unregistered]] unregistered <br> | [[Image:RegLib_C44_unregistered.gif|400px|original unregistered]] unregistered <br> | ||
| − | [[Image:RegLib_C44_registered_affine.gif|400px|registered (affine)]] registered (affine) | + | [[Image:RegLib_C44_registered_affine.gif|400px|registered (affine)]] registered (affine)<br> |
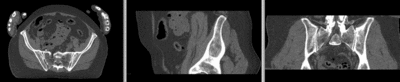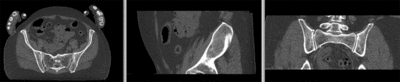
[[Image:RegLib_C44_registered_noMask.gif|400px|registered (nonrigid w/o masking)]] registered (nonrigid w/o masking)<br> | [[Image:RegLib_C44_registered_noMask.gif|400px|registered (nonrigid w/o masking)]] registered (nonrigid w/o masking)<br> | ||
[[Image:RegLib_C44_registered_withMask.gif|400px|registered (nonrigid+masking)]] registered (nonrigid+masking)<br> | [[Image:RegLib_C44_registered_withMask.gif|400px|registered (nonrigid+masking)]] registered (nonrigid+masking)<br> | ||
Latest revision as of 18:14, 1 October 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C44Contents
Slicer Registration Library Case 44: Visible Human Pelvis CT
Input
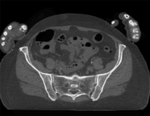
|
|

|
| baseline image | follow-up |
Modules used
Description
This dataset contains CT of the visible human male and female pelvis. This serves as a test example for exploring non-rigid registration for inter-subject comparison from CT. The overall strategy will be to register "vhf" to "vhm" via first affine and then BSpline registration. We will generate a mask to focus the registration on the bone structure only and ignore the soft tissue when computing the deformation. Because our original images are quite large (512x512x150), we will subsample the vhf pair for use with the Deformation Field Visualizer module, which might otherwise become too memory intensive.
Download
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C44.mrb (input data only, Slicer mrb file. 74 MB)
- RegLib_C44_full.mrb(input data + results, Slicer mrb file. 82 MB).
Video Screencasts
- Movie/screencast showing generating a registration mask
- Movie/screencast showing affine and nonrigid BSpline registration
- Movie/screencast showing visualization of the deformation via the Transform Visualizer module
Keywords
CT, pelvis, visible human, inter-subject
Procedure / Pipeline
- Mask generation: open the Editor module. Movie/screencast showing this step
- "Master Volume": select vhm
- A new labelmap "vhm-label" will be created
- Select "vhm" to be visible in the slice viewer
- Select the Threshold tool from the editor toolbar
- Adjust the lower threshold (slider bar) until most of the bone is highlighted,just before speckle noise starts to become included e.g. somewhere around an intensity value of 80. Leave the upper threshold unchanged at the max.
- Click Apply
- clean the segmentation:
- Select the "Identify Islands" editor effect.
 This will identify all continuous areas that are disconnected from each other. Click "Apply"
This will identify all continuous areas that are disconnected from each other. Click "Apply" - you should see the bones of the arms being assigned a different label value & color. We can now delete them with one click:
- select the "Change Island" effect
 . Change the label value to 0 (zero).
. Change the label value to 0 (zero). - in the axial (red) view, click the left mouse within the segmented areas of the arms.
- select the "Dilate" effect
 . Click the "Apply" button 3-4 times until the boundary of the segmentation extends well beyond the bone, including a several pixel wide layer of adjacent tissue.
. Click the "Apply" button 3-4 times until the boundary of the segmentation extends well beyond the bone, including a several pixel wide layer of adjacent tissue. - repeat the above for "vhf".
- save the two segmentations.
- Affine Registration: open the General Registration (BRAINS) module. Movie/screencast showing this step
- Fixed Image Volume: vhm
- Moving Volume: vhf
- check boxes for Include Rigd registr. phase , Include ScaleVersor3D, include Affine
- Slicer Linear Transform: select "create new transform", rename to "Xf1_Affine" or similar
- leave rest at defaults. Click Apply
- registration should take ~ 10 secs.
- use fade slider to verify alignment; compare with result snapshots shown below. Alignment will not be perfect but should be better than before.
- note: you can also change the colormaps for the fixed and moving volumes to better judge the alignment: go to the Volumes module and in the Display tab, select "green" and "magenta" as the respective colormaps for the two volumes (vhf, vhm)
- Nonrigid Registration (masked): open the General Registration (BRAINS). Movie/screencast showing this step
- Fixed Image Volume: vhm Moving: vhf
- Registration phases: from Initialize with previously generated transform', select "Xf1_Affine" node created before.
- Registration phases: uncheck boxes for rigid, scale and affine and check box for BSpline
- Output: Slicer Linear transform: set to None
- Output: Slicer BSpline transform: create new, rename to "Xf2_BSpline_msk" or similar
- Output Image Volume: create new, rename to "vhf_Xf2"; Pixel Type: "short"
- Registration Parameters: increase Number Of Samples to 200,000; Number of Grid Subdivisions: 7,7,7
- Control Of Mask Processing Tab: check ROI box, for Input Fixed Mask and Input Moving Mask select the two dilated labelmaps from above
- Leave all other settings at default
- click apply
- Deformation Visualization: Movie/screencast showing this step
- if you have not yet installed the Deformation Field Visualizer extension, see here for a movie clip on how to install it.
- we first generate a smaller version of our image to save memory:
- open the Resize Image (BRAINS) module (under Registration)
- Image To Warp: select "vhf"
- Output Image: create new, rename to "vhf_small" or similar
- Pixel Type: select "short"
- Scale Factor: leave at 2.0
- click Apply
- open the Transform Visualizer module (under: All Modules)
- Deformation: select "Xf2_BSpline"
- Reference Image: select "vhf_small" generated above
- Visualization mode: click on "Grid Slice"
- Grid Slice Options: Slice: red; Spacing: 20mm
- Click Apply. you should see the deformation field overlay. Adjust slice, spacing etc. to taste.
Registration Results
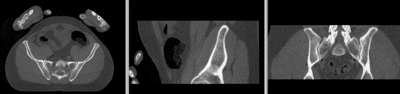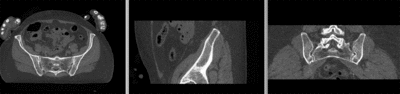 unregistered
unregistered
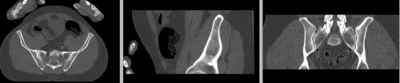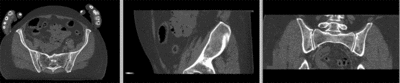 registered (affine)
registered (affine)
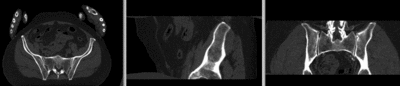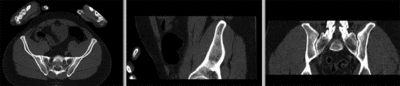 registered (nonrigid w/o masking)
registered (nonrigid w/o masking)
 registered (nonrigid+masking)
registered (nonrigid+masking)
 registered deformation only of vhf
registered deformation only of vhf
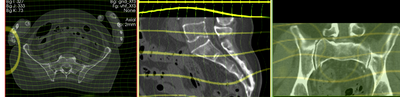 deformation visualized by grid image overlay
deformation visualized by grid image overlay
Acknowledgments
Original CT from the Visible Human Project shared by the University of Iowa.