Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C32"
| (11 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
== Input == | == Input == | ||
{| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image:RegLib_C32_thumb3.png| | + | |[[Image:RegLib_C32_thumb3.png|250px|lleft|this is the fixed reference image. All images are aligned into this space]] |
| − | |[[Image:RegLib_C32_thumb2.png| | + | |[[Image:RegLib_C32_thumb2.png|250px|lleft|this is the fixed reference image. All images are aligned into this space]] |
|[[Image:RegArrow_NonRigid.png|100px|lleft]] | |[[Image:RegArrow_NonRigid.png|100px|lleft]] | ||
|[[Image:RegArrow_Affine.png|100px|lleft]] | |[[Image:RegArrow_Affine.png|100px|lleft]] | ||
| Line 17: | Line 17: | ||
|moving image<br>T1 SPGR | |moving image<br>T1 SPGR | ||
|} | |} | ||
| + | |||
| + | == Description == | ||
| + | This is an example of the inverted workflow, i.e. we seek to align the structural T1 to the DTI. <br> | ||
| + | '''Approach''': to align the DTI and the T1 we need 2 preprocessing steps: 1. reduce the bias field inhomogeneity in the reference T1 and 2. obtain a skull-stripping / brain mask for the T1. Since the DWI is already and will not move, no resampling is required before obtaining the DTI (DTI estimation). On instructions on how to generate the DTI from the DWI see, for example [[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C29|'''Case 29''']]. As common for DTI, the T1-DTI registration will include non-rigid deformation to correct for the strong distortions from the EPI acquisition. | ||
== Modules used == | == Modules used == | ||
| Line 22: | Line 26: | ||
*[[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | *[[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
== Download (from NAMIC MIDAS) == | == Download (from NAMIC MIDAS) == | ||
<small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> | ||
| − | *[http://slicer.kitware.com/midas3/download/?items= | + | *[http://slicer.kitware.com/midas3/download/?items=103594 '''RegLib_C32.mrb'''] <small>(input data, Slicer mrb file. 31 MB). </small> <br> |
| − | *[http://slicer.kitware.com/midas3/download/?items= | + | *[http://slicer.kitware.com/midas3/download/?items=103593 '''RegLib_C32_full.mrb'''] <small>(input data+results, Slicer mrb file. 36 MB). </small> |
== Keywords == | == Keywords == | ||
MRI, brain, head, intra-subject, DTI, T1, T2, non-rigid, tumor, surgical planning | MRI, brain, head, intra-subject, DTI, T1, T2, non-rigid, tumor, surgical planning | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== Video Screencasts === | === Video Screencasts === | ||
| Line 94: | Line 83: | ||
== Registration Results== | == Registration Results== | ||
{|cellpadding="10" cellspacing="0" border="0" | {|cellpadding="10" cellspacing="0" border="0" | ||
| − | |[[Image: | + | |[[Image:RegLib_C32_unregistered.png|400px]] ||unregistered pair of T1 and DTI (click to enlarge)<br> |
| + | |- | ||
| + | |[[Image:RegLib_C32_RegisteredAffine.gif|400px]] || T1 registered to DTI baseline : Affine only(click to enlarge) | ||
| + | |- | ||
| + | |[[Image:RegLib_C32_registered.gif|400px]] || T1 registered to DTI Affine + BSpline (click to enlarge) | ||
|} | |} | ||
Latest revision as of 13:46, 3 September 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C32
|
For the latest Slicer documentation, visit the read-the-docs. |
Contents
Slicer Registration Library Case #29: Intra-subject Brain DTI
Input

|
|
|
|
|
| fixed image DTI |
fixed image DTI baseline |
moving image T1 SPGR |
Description
This is an example of the inverted workflow, i.e. we seek to align the structural T1 to the DTI.
Approach: to align the DTI and the T1 we need 2 preprocessing steps: 1. reduce the bias field inhomogeneity in the reference T1 and 2. obtain a skull-stripping / brain mask for the T1. Since the DWI is already and will not move, no resampling is required before obtaining the DTI (DTI estimation). On instructions on how to generate the DTI from the DWI see, for example Case 29. As common for DTI, the T1-DTI registration will include non-rigid deformation to correct for the strong distortions from the EPI acquisition.
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C32.mrb (input data, Slicer mrb file. 31 MB).
- RegLib_C32_full.mrb (input data+results, Slicer mrb file. 36 MB).
Keywords
MRI, brain, head, intra-subject, DTI, T1, T2, non-rigid, tumor, surgical planning
Video Screencasts
- Phase 1-3: registering T1 to a DTI image (5 min, 22MB)
Procedures
- Phase 1: Preprocessing: Bias Correction of T1
- open the N4ITKBiasFieldCorrection module
- Input image: T1
- Output Volume: create & rename new, rename to "T1_biascorr" or similar
- Leave all other settings at defaults. Click: Apply. Processing may take 1-2 minutes based on your CPU.
- Phase 2: Affine Registration
We first perform an affine registration for a rough alignment. Once this succeeds we continue to the nonrigid (BSpline) portion. Because Affine registration does not require resampling, there is no cost (in terms of data loss) of separating this step.
- open the General Registration (BRAINS) module
- Fixed Image Volume: DTI_baseline
- Moving Image Volume: T1_biascorr
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create & rename new transform, rename to "Xf1_DTI-T1"
- Output Image Volume: create new volume, rename to T1_Xf1 (we use this only to verify the result, we can discard afterwards)
- Registration Phases: check boxes for Rigid, Rigid+Scale, Affine
- Main Parameters:
- Number Of Samples: 200,000
- Leave all other settings at default
- click: Apply.
- upon completion: place T1_Xf1 and DTI_baseline into foreground and background, respectively, and fade back and forth to verify alignment. Areas of DTI distortions need not match up exactly, but the overall parenchyma should be well aligned.
- Phase 3: BSpline Registration
We now proceed to the nonrigid (BSpline) portion, using the above Affine transform as a starting point. This creates a new transform that includes both the affine and the BSpline portion. Because nonrigid transforms cannot be visualized directly, a resampled volume is required for this step.
- open the General Registration (BRAINS) module
- Fixed Image Volume: DTI_baseline
- Moving Image Volume: T1_biascorr (not the T1_Xf1)
- Output Settings:
- Slicer BSpline Transform": create & rename new transform, rename to "Xf2_BSpline"
- Slicer Linear Transform: none
- Output Image Volume: create & rename new volume, rename to T1_Xf2
- Registration Phases: uncheck boxes for Rigid, Rigid+Scale, Affine , check box for BSpline
- Main Parameters:
- Number Of Samples: increase to 300,000 (or more)
- B-Spline Grid Size: 3,3,3
- Mask Option: select ROI button
- Leave all other settings at default
- click: Apply. This may take 1-2 minutes depending on your CPU.
- as above, verify by fading between DTI_baseline and T1_Xf2
- you can also examine the deformation portion only by placing T1_Xf1 and T1_Xf2 into fore- and background and fading between them.
- Save results.
Registration Results
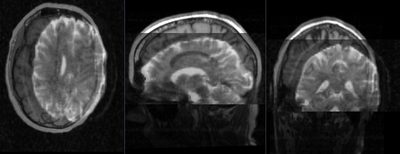 |
unregistered pair of T1 and DTI (click to enlarge) |
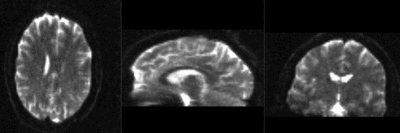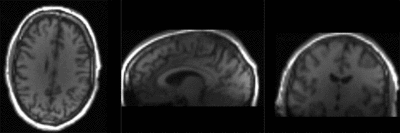 |
T1 registered to DTI baseline : Affine only(click to enlarge) |
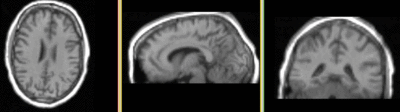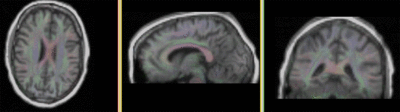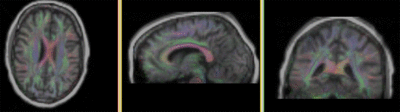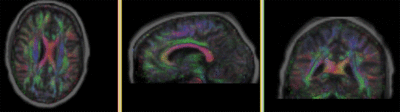 |
T1 registered to DTI Affine + BSpline (click to enlarge) |