Documentation:Nightly:Registration:RegistrationLibrary:RegLib C27
Contents
Slicer Registration Library Case #27: Intra-subject Brain DTI
Input
|
|
|
|
|
|
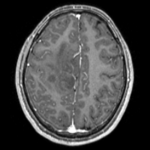
| moving image T1 SPGR |
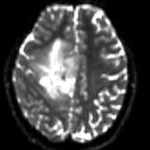
fixed image 1 T2 |
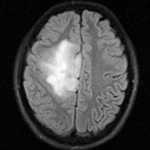
moving image DTI baseline |
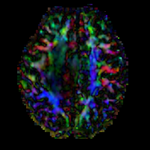
moving image DTI tensor |
Description
This is a classic case of a multi-sequence MRI exam we wish to spatially align to the anatomical reference scan (T1-SPGR). The scan of interest is the DTI image to be aligned for surgical planning/reference. The key points are: 1) the DWI sequence (EPI) contains distortions we seek to correct via non-rigid alignment; 2) the DTI baseline is similar in contrast to a T2, albeit at much lower resolution, but we do not have a T2 weighted anatomical scan, but instead a T1 and a FLAIR. So we use the FLAIR as the fixed image, because its contrast matches the DTI-baseline better than the T1, esp. in terms of tumor contrast.
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C27.mrb (input data, Slicer mrb file 50MB)
- RegLib_C27_full.mrb (input data + results, Slicer mrb file 114MB)
Keywords
MRI, brain, head, intra-subject, DTI, T1, T2, non-rigid, tumor, surgical planning
Registration Approach
- a direct registration of the DTI_baseline to the T1 is not recommended, since the FLAIR is a better match in contrast to the DTI baseline. We therefore first align the DTI with the FLAIR and then also register the T1 to the FLAIR. This minimizes resampling to only 1 image and only 1 transform.
- the DTI-FLAIR registration includes non-rigid deformation to correct for the strong distortions from the EPI acquisition. Because of the nonrigid component a mask of the brain parenchyma helps in obtaining a meaningful transform. Registration without a mask is possible but not as good as with a mask.
Video Screencasts
- Phase 1:Register DTI baseline to FLAIR
- Phase 2:Affine Register the T1 to the FLAIR
- Phase 3:Resample the DTI with the obtained transform
Procedures
- Phase 1: register DTI
- open the General Registration (BRAINS) module
- Fixed Image Volume: FLAIR
- Moving Image Volume: DTI_baseline
- Output Settings:
- Slicer BSpline Transform": create & rename new transform, rename to "Xf1_DTI-FLAIR"
- Slicer Linear Transform: none
- Output Image Volume: create new volume, rename to DTI_baseline_Xf1
- Registration Phases: check boxes for Rigid, Rigid+Scale, Affine and BSpline
- Main Parameters:
- Number Of Samples: 300,000
- B-Spline Grid Size: 7,7,5
- Leave all other settings at default
- click: Apply. This may take several minutes depending on your CPU.
- Save Results
- Phase 2: register T1 to FLAIR (without resampling)
- open the General Registration (BRAINS) module
- Fixed Image Volume: FLAIR
- Moving Image Volume: T1
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create & rename new transform, rename to "Xf2_T1-FLAIR"
- Output Image Volume: none (since this is an affine transform we need not resample to see the result)
- Registration Phases: check boxes for Rigid and Affine
- Main Parameters : Number Of Samples: 200,000
- Leave all other settings at default
- click: Apply; runtime < 10 sec (MacPro QuadCore 2.4GHz) -> This only generates the transform. We still need to apply it to the image
- Apply the transform to the T1 image: open the Data module
- locate the transform generated above "Xf2_T1-FLAIR" and the T1 image. Drag the T1 image node onto the transform. A little + sign should appear next to the transform. Click on it to verify the T1 image was placed under the transform.
- right-click on the T1 image node, from the pop-up menu select Harden Transform. The T1 node moves back out to the main level
- double-click on the T1 image node, rename it to "T1_Xf2" or similar to document that this image is now in a new orientation (no longer the original T1). Note that when you save the new T1 it will not resample the image data, but rather write its new orientation as part of the image header. If you open the image with software that ignores this header information, it will look the same as the original T1. Also note that when you save your result, the save dialog may still display the name of the original T1 file. Make sure to change the filename before saving to prevent overwriting the original.
- Save Results
- Phase 3: resample DTI
- Open the Resample DTI Volume module] (under All Modules menu; note this is distinct from the ResampleScalarVectorDWIVolume). We must use the ResampleDTI module to process tensor images to correctly resample the tensor data.
- Input Volume: DTI
- Output Volume: create new DTI Volume, rename to DTI_Xf2
- Reference Volume: T2_Xf1
- Transform Node: select "Xf2_DTI-T1" created above
- check box: displacement
- leave all other settings at defaults
- Click Apply; runtime ~ 3 min.
- set T1 or FLAIR as background and the new DTI_Xf2 volume as foreground
- Move fade slider to see DTI overlay onto the structural image
- Save Results
Registration Results
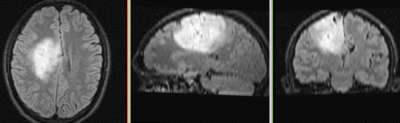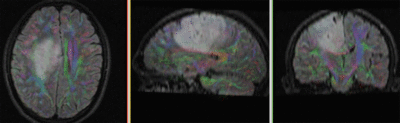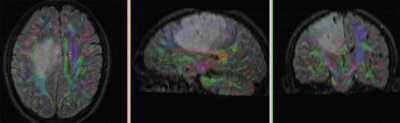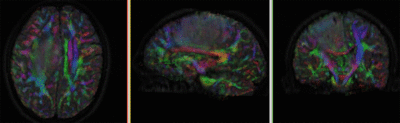 |
original and registered pair of FLAIR and DTI (click to enlarge) |
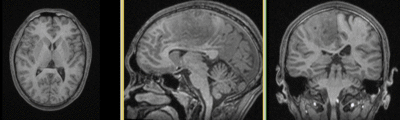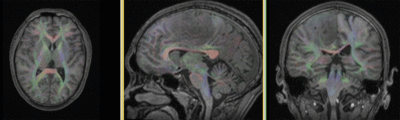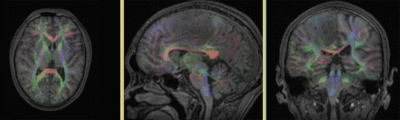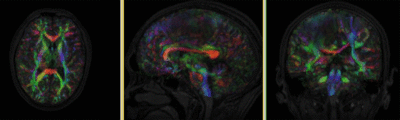 |
original and registered pair of T1 and DTI (click to enlarge) |