Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C21"
From Slicer Wiki
(Created page with 'Back to Registration Library <br> = Slicer Registration Library Case #21: Align knee MRI of two different subjects = =…') |
|||
| Line 26: | Line 26: | ||
#'''Compute Rigid Registration''': open the [[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | #'''Compute Rigid Registration''': open the [[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | ||
##''Input Images: Fixed Image Volume'': MRI_58 | ##''Input Images: Fixed Image Volume'': MRI_58 | ||
| − | ## | + | ##''Input Images: Moving Image Volume'': MRI_64 |
##''Output Settings'': | ##''Output Settings'': | ||
| − | ###''Slicer Linear Transform'' (create new transform, rename to: "Xf1_Rigid") | + | ###''Slicer Linear Transform'': (create new transform, rename to: "Xf1_Rigid") |
| − | ###''Output Image Volume'' none (we need no resampled volume at this stage, only the transform) | + | ###''Output Image Volume'': none (we need no resampled volume at this stage, only the transform) |
##''Registration Phases'': select/check ''Rigid'' | ##''Registration Phases'': select/check ''Rigid'' | ||
##''Main Parameters'': | ##''Main Parameters'': | ||
| Line 37: | Line 37: | ||
##check the result | ##check the result | ||
###Place "MRI_58" in the background and "MRI_64" in the foreground | ###Place "MRI_58" in the background and "MRI_64" in the foreground | ||
| − | ###switch to the [[Documentation/Nightly/Modules/Data|''Data'' module]], locate the "MRI_64" node, and drag it onto the newly created " | + | ###switch to the [[Documentation/Nightly/Modules/Data|''Data'' module]], locate the "MRI_64" node, and drag it onto the newly created "Xf1_Rigid" node. |
###fade between background and foreground to judge the alignment. | ###fade between background and foreground to judge the alignment. | ||
| − | ###for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the [[Documentation/Nightly/Modules/Volumes|''Volumes'' #'''Compute Affine Registration''': return to the [[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | + | ###for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the [[Documentation/Nightly/Modules/Volumes|''Volumes'' module]], select "MRI_58" and then select "Green" from the ''Lookup Table'' menu. Similarly select "Cyan" for "MRI_64". |
| + | #'''Compute Affine Registration''': return to the [[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] | ||
##''Input Images: Fixed Image Volume'': MRI_58 | ##''Input Images: Fixed Image Volume'': MRI_58 | ||
| − | ## | + | ##''Input Images: Moving Image Volume'': MRI_64 |
##''Output Settings'': | ##''Output Settings'': | ||
| − | ###''Slicer Linear Transform'' (create new transform, rename to: "Xf2_Affine") | + | ###''Slicer Linear Transform'': (create new transform, rename to: "Xf2_Affine") |
| − | ###''Output Image Volume'' none (we need no resampled volume at this stage, only the transform) | + | ###''Output Image Volume'': none (we need no resampled volume at this stage, only the transform) |
##''Initialization'': for the ''Initialization transform'' select the "Xf1_Rigid" transform created above. | ##''Initialization'': for the ''Initialization transform'' select the "Xf1_Rigid" transform created above. | ||
##''Initialize Transform Mode'': switch to ''Off'' | ##''Initialize Transform Mode'': switch to ''Off'' | ||
| − | ##''Registration Phases'': '''un'''check ''Rigid'' and select ''Rigid+Scale | + | ##''Registration Phases'': '''un'''check ''Rigid'' and select ''Rigid+Scale'' and ''Affine'' only |
##''Main Parameters'': | ##''Main Parameters'': | ||
###leave ''Number Of Samples'' to 200,000 | ###leave ''Number Of Samples'' to 200,000 | ||
| Line 57: | Line 58: | ||
###fade between background and foreground to judge the alignment. | ###fade between background and foreground to judge the alignment. | ||
###for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the [[Documentation/Nightly/Modules/Volumes|''Volumes'' module]], select "MRI_58" and then select "Green" from the ''Lookup Table'' menu. Similarly select "Cyan" for "MRI_64". | ###for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the [[Documentation/Nightly/Modules/Volumes|''Volumes'' module]], select "MRI_58" and then select "Green" from the ''Lookup Table'' menu. Similarly select "Cyan" for "MRI_64". | ||
| − | #''' | + | #'''Compute BSpline Registration''': return to the [[Documentation/Nightly/Modules/BRAINSFit|''General Registration (BRAINS)'' module]] |
| − | + | ##''Input Images: Fixed Image Volume'': MRI_58 | |
| − | ### | + | ##''Input Images: Moving Image Volume'': MRI_64 |
| − | ### | + | ##''Output Settings'': |
| − | ###apply the transform | + | ###''Slicer BSpline Transform'': Create and rename new. Rename to "MRI_64_Xf3" |
| − | + | ###''Slicer Linear Transform'': set to ''none'' | |
| − | ## | + | ###''Output Image Volume'': Create and rename new. Rename to "MRI_64_Xf3" |
| + | ##''Initialization'': for the ''Initialization transform'' select the "Xf1_Affine" transform created above. | ||
| + | ##''Initialize Transform Mode'': switch to ''Off'' | ||
| + | ##''Registration Phases'': '''un'''check ''Rigid+Scale'' and ''Affine'' and select ''BSpline'' only | ||
| + | ##''Main Parameters'': | ||
| + | ###increase ''Number Of Samples'' to 400,000 | ||
| + | ###set ''B-Spline Grid Size'' to 5,5,3 | ||
| + | ##Leave all other settings at default | ||
| + | ##click: ''Apply''; runtime for this will be more than 1 minute, depending on CPU. | ||
| + | ##check the result | ||
| + | ###Place "MRI_58" in the background and "MRI_64_Xf3" in the foreground | ||
| + | #Apply and save transforms | ||
| + | ##To apply the rigid and affine transforms you need not resample the volume. Simply drag the MRI_64 volume into the transform node (Xf1_Rigid or Xf2_Affine), and then right click on the "MRI_64" volume node and from the pop-up menu, select ''Harden Transform''. This will apply the transform '''without resampling''' and move it back to the main hierarchy level. '''Note that now you have a reoriented volume although the name is the same. It is advisable to immediately rename the node to document this fact. Then save it as a new file under a new name.''' | ||
| + | ##To apply the nonrigid transform resampling is required. You already have a resampled volume "MRI_64_Xf3" from above. This uses linear interpolation and float as default. You can change these settings under the ''Advanced Output Settings'' tab. For this example "short" as Pixel Type and "Windowed Sinc" is recommended. To obtain a new resampled copy without repeating the whole registration step, use the [[Documentation/Nightly/Modules/BRAINSResample|''BRAINSResample'' module]] | ||
==Registration Results== | ==Registration Results== | ||
Revision as of 15:28, 19 August 2013
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C21Contents
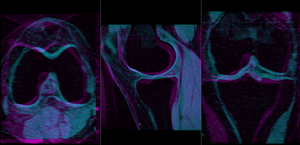
Slicer Registration Library Case #21: Align knee MRI of two different subjects
Input
|
lleft | 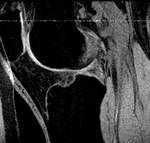
|
| fixed image/target MRI |
moving image MRI |
Modules used
Download (from NAMIC MIDAS)
Keywords
MRI, knee, inter-subject, nonrigid
Procedure
It is possible to perform all 3 steps of rigid, affine and BSpline registration in a single pass within the General Registration (BRAINS) module, but to better show/judge progress we perform each separately and feed the results of one the the next step. Hence we first run a rigid registration, then use that to compute an affine registration, and finally use the affine as starting point for a BSpline registration. For the last one we will need to generate a quick mask to prevent aliasing artifacts in the image from influencing the result. The masks are available in the download set.
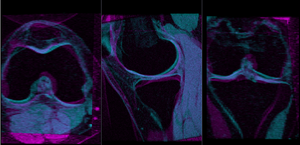
- Compute Rigid Registration: open the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: MRI_58
- Input Images: Moving Image Volume: MRI_64
- Output Settings:
- Slicer Linear Transform: (create new transform, rename to: "Xf1_Rigid")
- Output Image Volume: none (we need no resampled volume at this stage, only the transform)
- Registration Phases: select/check Rigid
- Main Parameters:
- increase Number Of Samples to 200,000
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- check the result
- Place "MRI_58" in the background and "MRI_64" in the foreground
- switch to the Data module, locate the "MRI_64" node, and drag it onto the newly created "Xf1_Rigid" node.
- fade between background and foreground to judge the alignment.
- for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the Volumes module, select "MRI_58" and then select "Green" from the Lookup Table menu. Similarly select "Cyan" for "MRI_64".
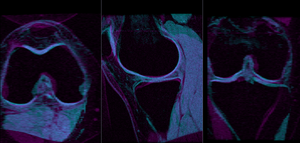
- Compute Affine Registration: return to the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: MRI_58
- Input Images: Moving Image Volume: MRI_64
- Output Settings:
- Slicer Linear Transform: (create new transform, rename to: "Xf2_Affine")
- Output Image Volume: none (we need no resampled volume at this stage, only the transform)
- Initialization: for the Initialization transform select the "Xf1_Rigid" transform created above.
- Initialize Transform Mode: switch to Off
- Registration Phases: uncheck Rigid and select Rigid+Scale and Affine only
- Main Parameters:
- leave Number Of Samples to 200,000
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- check the result
- Place "MRI_58" in the background and "MRI_64" in the foreground
- switch to the Data module, locate the "MRI_64" node, and drag it onto the newly created "Xf1_Affine" node.
- fade between background and foreground to judge the alignment.
- for better visibility, you can change the colormaps of the two volumes, as used in the examples shown here. Switch to the Volumes module, select "MRI_58" and then select "Green" from the Lookup Table menu. Similarly select "Cyan" for "MRI_64".
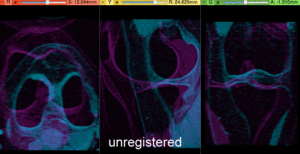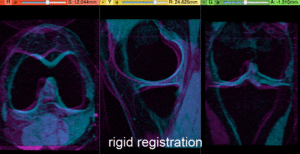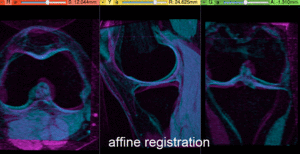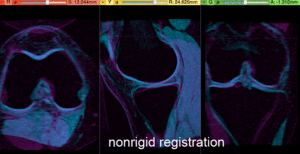
- Compute BSpline Registration: return to the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: MRI_58
- Input Images: Moving Image Volume: MRI_64
- Output Settings:
- Slicer BSpline Transform: Create and rename new. Rename to "MRI_64_Xf3"
- Slicer Linear Transform: set to none
- Output Image Volume: Create and rename new. Rename to "MRI_64_Xf3"
- Initialization: for the Initialization transform select the "Xf1_Affine" transform created above.
- Initialize Transform Mode: switch to Off
- Registration Phases: uncheck Rigid+Scale and Affine and select BSpline only
- Main Parameters:
- increase Number Of Samples to 400,000
- set B-Spline Grid Size to 5,5,3
- Leave all other settings at default
- click: Apply; runtime for this will be more than 1 minute, depending on CPU.
- check the result
- Place "MRI_58" in the background and "MRI_64_Xf3" in the foreground
- Apply and save transforms
- To apply the rigid and affine transforms you need not resample the volume. Simply drag the MRI_64 volume into the transform node (Xf1_Rigid or Xf2_Affine), and then right click on the "MRI_64" volume node and from the pop-up menu, select Harden Transform. This will apply the transform without resampling and move it back to the main hierarchy level. Note that now you have a reoriented volume although the name is the same. It is advisable to immediately rename the node to document this fact. Then save it as a new file under a new name.
- To apply the nonrigid transform resampling is required. You already have a resampled volume "MRI_64_Xf3" from above. This uses linear interpolation and float as default. You can change these settings under the Advanced Output Settings tab. For this example "short" as Pixel Type and "Windowed Sinc" is recommended. To obtain a new resampled copy without repeating the whole registration step, use the BRAINSResample module
Registration Results