|
|
| Line 15: |
Line 15: |
| | * [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] | | * [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module] |
| | | | |
| − | ===Download ===
| + | == Download (from NAMIC MIDAS) == |
| − | *[[Media:RegLib_C12_Data.zip|'''download input image data''' <small> (Input Data, Masks, Solution Transforms, Resampled results (NRRD images, zip file 65 MB) </small>]] | + | <small>''Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself. ''</small> |
| | + | *[http://slicer.kitware.com/midas3/download/?items=95490 '''RegLib_C03.mrb''': input data only, use this to run the tutorial from the start <small>(Slicer mrb file. 50 MB). </small>] |
| | + | *[http://slicer.kitware.com/midas3/download/?items=95491 '''RegLib_C03_full.mrb''': includes raw data + all solutions and intermediate files, use to browse/verify <small>(Slicer mrb file. 108 MB). </small>] |
| | | | |
| | ===Objective / Background === | | ===Objective / Background === |
| Line 26: |
Line 28: |
| | ===Input Data=== | | ===Input Data=== |
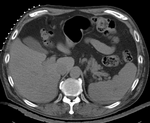
| | *reference/fixed : pr-op CT, 0.95 x 0.95 x 5 mm voxel size | | *reference/fixed : pr-op CT, 0.95 x 0.95 x 5 mm voxel size |
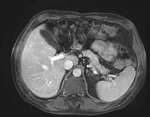
| − | *moving: intra-op MRI, 0.78 x 0.78 x 2.5 mm axial, | + | *moving: intra-op MRI, 0.78 x 0.78 x 2.5 mm axial, |
| − | | |
| − | === Discussion: Registration Challenges ===
| |
| − | *large differences in FOV
| |
| − | *strong differences in image contrast between MRI & CT
| |
| − | *contrast enhancement and pathology and treatment changes cause additional differences in image content
| |
| − | *we have strongly anisotropic voxel sizes with much less through-plane resolution
| |
| − | | |
| − | === Notes / Overall Strategy ===
| |
| − | *masking is required to focus the registration algorithm on the structure of interest
| |
| − | *Overall strategy:
| |
| − | :#obtain (manual) a coarse segmentation of the liver in both MRI and CT. Dilate by a few pixels to include the organ boundary
| |
| − | :#perform a manual initial alignment of MR to CT. Use this alignment as starting point for the automated registration
| |
| − | :#run an affine registration with above masks and intial alignment
| |
| − | #run a non-rigid BSpline registration with above affine alignment as starting point
| |
| − | | |
| − | | |
| − | === Procedures ===
| |
| − | *'''Phase I: Build Masks'''
| |
| − | : Note: for illustration the example set contains 2 masks: one with only the liver and one also including spleen and kidney (Mask2). As shown in the results below, the liver-only mask is insufficiently constraining the registration, yielding a result that at first glance looks ok for the liver, but has significant misalignment in the remaining abdominal area. Hence it is advisable to stabilize the registration further by including more structures with good contrast in both images (Spleen, Kidney).
| |
| − | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/Editor Editor module]
| |
| − | #select "MRI" as the master volume ; a new "MRI_label" is created
| |
| − | #using the Brush tool, trace the liver contour from axial slices. Also include kidneys and spleen.
| |
| − | #repeat the same for the CT.
| |
| − | #you should end up with 2 label maps similar to those included in the example dataset. See snapshots in Result section below.
| |
| − | #save the label maps as "MRI-label.nrrd" or similar
| |
| − | #registration masks ideally extend beyond the structure boundary:
| |
| − | #select the ''Dilate'' tool and click ''Apply'' 3-4 times to extend the mask area
| |
| − | #repeat for both masks
| |
| − | #In the [http://www.slicer.org/slicerWiki/index.php/Modules:Editor-Documentation-3.6 ''Editor'' module], use the ''Dilate'' function to expand the outline by 2-3 pixels (click on ''Apply'' button 2-3 times)
| |
| − | #save dilated labelmasks under new name (e.g. CT_mask.nrrd)
| |
| − | *'''Phase II: MR-CTpre registration''' (Affine)
| |
| − | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module]
| |
| − | ##''Fixed Image Volume'': CT
| |
| − | ##''Moving Image Volume'': MRI
| |
| − | ##Output Settings:
| |
| − | ###''Slicer BSpline Transform": none
| |
| − | ###''Slicer Linear Transform'': create new transform, rename to "Xf1_MRI-CT_Affine"
| |
| − | ###''Output Image Volume'': create new volume, rename to "MRI_Xf1" (we use this for ease of validation only)
| |
| − | ##''Initialization'': select the ''useCenterOfROIAlign''
| |
| − | ##''Registration Phases'': check boxes for ''Rigid'' , ''Rigid+Scale'' and ''Affine''
| |
| − | ##''Mask Option'': select ''ROI'' button
| |
| − | ###''(ROI)Masking input fixed'': CT_mask
| |
| − | ###''(ROI)Masking input moving'': MRI_mask
| |
| − | ##Leave all other settings at default
| |
| − | ##click: Apply; runtime < 1 min (MacPro QuadCore 2.4GHz)
| |
| − | ##this should generate a first alignment.
| |
| − | *'''Phase III: Nonrigid Registration'''
| |
| − | #open the [http://wiki.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/BRAINSFit General Registration (BRAINS) module]
| |
| − | ##''Fixed Image Volume'': CT
| |
| − | ##''Moving Image Volume'': MRI
| |
| − | ##Output Settings:
| |
| − | ###''Slicer BSpline Transform": create new transform, rename to "Xf2_MRI-CT_BSpline"
| |
| − | ###''Slicer Linear Transform'': none
| |
| − | ###''Output Image Volume'': create new volume, rename to "MRI_Xf2"
| |
| − | ##''Initialization'':
| |
| − | ###''Initialization Transform'': select "Xf1_MRI-CT_Affine" created in Phase II above
| |
| − | ###''Initialization Transform Mode'': Off
| |
| − | ##''Registration Phases'': check boxes for ''BSpline'' only
| |
| − | ##''Main Parameters'':
| |
| − | ###''Number Of Samples'': 200,000
| |
| − | ###''B-Spline Grid Size'': 7,7,3
| |
| − | ##''Mask Option'': select ''ROI'' button
| |
| − | ###''(ROI)Masking input fixed'': CT_mask
| |
| − | ###''(ROI)Masking input moving'': MRI_mask
| |
| − | ##Leave all other settings at default
| |
| − | ##click: Apply
| |
| − | | |
| − | === Registration Results===
| |
| − | {| style="color:#bbbbbb; " cellpadding="10" cellspacing="0" border="0"
| |
| − | |[[Image:RegLib_C12_unregistered.gif|300px|left|unregistered MRI & CT]]
| |
| − | |unregistered MRI & CT
| |
| − | |-
| |
| − | |[[Image:RegLib_C12_Masks.png|300px|left|registration masks]]
| |
| − | |registration masks
| |
| − | |-
| |
| − | |[[Image:RegLib_C12_Affine.gif|300px|left|affine registered MRI & CT]]
| |
| − | |affine registered MRI & CT
| |
| − | |-
| |
| − | |[[Image:RegLib_C12_BSpline.gif|300px|left|nonrigid registered MRI & CT]]
| |
| − | |nonrigid registered MRI & CT
| |
| − | |-
| |
| − | |[[Image:RegLib_C12_ConstrainFailure.gif|300px|left|insufficient mask: nonrigid registered with liver mask only, note the misalignment in the remaining abdomen]]
| |
| − | |insufficient mask: nonrigid registered with liver mask only, note the misalignment in the remaining abdomen
| |
| − | |-
| |
| − | |[[Image:RegLib_C12_ColorOverlay.gif|300px|left|Color overlay of registered MRI onto CT, illustrating the fusion: MRI soft tissue contrast shows substructures & vasculature invisible on the CT]]
| |
| − | |Color overlay of registered MRI onto CT, illustrating the fusion: MRI soft tissue contrast shows substructures & vasculature invisible on the CT
| |
| − | |}
| |
| − | | |
| − | | |
| − | === Acknowledgments ===
| |
| − | Thanks to Dr.Stuart Silverman and Dr. Nobuhiko Hata for sharing this case.
| |