Documentation:Nightly:Registration:RegistrationLibrary:RegLib C10
Contents
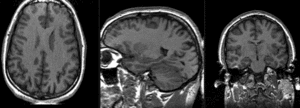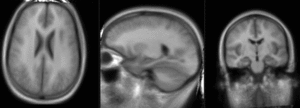
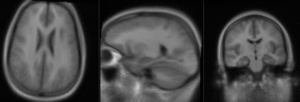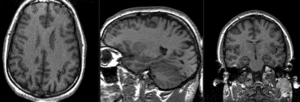
Slicer Registration Library Case #10: group atlas to structural MRI
Input
| fixed image/target T1 |
moving image group atlas |
Description
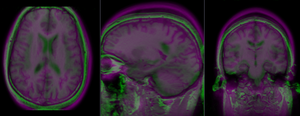
This is an example of inter-subject registration; we seek to align a group atlas image with a structural reference T1. Approach: we first compute an affine alignment of the atlas to the T1. We then proceed to nonrigid registrations of higher and higher DOF, stopping at the DOF where deformations become unreasonable. We can constrain the deformation by either explicit limits on the absolute deformation amount or via a mask. Constraints become increasingly critical as DOF increases.
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C10.mrb: input data only, use this to run the tutorial from the start (Slicer mrb file. 12 MB).
- RegLib_C10_full.mrb: includes raw data + all solutions and intermediate files, use to browse/verify (Slicer mrb file. 50 MB).
Keywords
MRI, atlas, brain, head, inter-subject
Video Screencasts
Procedure
- Affine Registration: register atlas to T1
- open the General Registration (BRAINS) module
- Fixed Image Volume: T1ref
- Moving Image Volume: Atlas
- Output Settings:
- Slicer BSpline Transform": none
- Slicer Linear Transform: create & rename new transform: Xf1_Affine.tfm
- Output Image Volume: none
- Registration Phases: check box for "Rigid"', "Rigid+Scale" and "Affine"
- Main Parameters
- Number of Samples: 200,000
- click: Apply; runtime < 10 sec (MacPro QuadCore 2.4GHz)
- Nonrigid BSpline Registration
- open the General Registration (BRAINS) module
- Fixed Image Volume: T1ref
- Moving Image Volume: Atlas
- Slicer BSpline Transform": create & rename new transform: Xf2_BSpline.tfm
- Slicer Linear Transform: none
- Output Image Volume: create & rename new: Atlas_Xf2
- Initialization Transform:select "Xf1_Affine" created above
- Initialization Transform Mode: off
- Registration Phases: uncheck boxe for Rigid and instead check BSpline
- Main Parameters: Number of Samples: 300,000
- BSpline Grid Size:5,5,3
- click: Apply
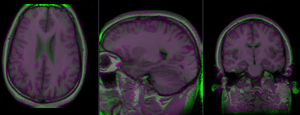
- Increasing DOF
- repeat the above with a larger BSpline grid:
- Slicer BSpline Transform": create & rename new transform: Xf3_BSpline2.tfm
- Output Image Volume: create & rename new: Atlas_Xf3
- Initialization Transform:select "Xf1_Affine" created above
- Initialization Transform Mode: off
- Main Parameters: Number of Samples: 400,000
- BSpline Grid Size:7,7,5
- click: Apply
- repeat the above with a larger BSpline grid:
- further increase DOF: you can continue exploring how far the direct approach (from Affine direct to high DOF BSpline) remains stable, by increasing the BSpline grid size until results become too distorted. Alternatively you can use the lower DOF result (e.g. Atlas_Xf2) as input to the next higher DOF transform. This enables a stable progression to higher DOF, but it also includes multiple resampling and accompanied data loss from interpolation blurring.