Documentation:Nightly:Registration:RegistrationLibrary:RegLib C04
Contents
Slicer Registration Library Case #2: Multi contrast MRI + segmentation labelmap
Input
| this is the main fixed reference image. All images are ev. aligned into this space | 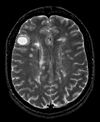
|
|
|
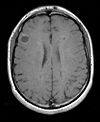
| exam 1: PD | exam 1: T2 | exam 1: T1-Gd | |

|
|||
|
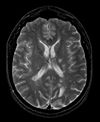
|

|
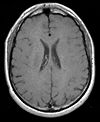
|
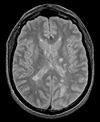
| exam 2: PD | exam 2: T2 | exam 2: T1-Gd |
Objective / Background
We have a structural T1-weighted MRI scan and a lesion segmentation obtained from another (FLAIR) scan. Goal is to align the FLAIR image and its segmentation labelmap with the structural reference T1 scan.
Approach: we first compute the registration from the T1-FLAIR pair and then apply this transform to the lesion segmentation.
Modules used
Download (from NAMIC MIDAS)
Why 2 sets of files? The "input data" mrb includes only the unregistered data to try the method yourself from start to finish. The full dataset includes intermediate files and results (transforms, resampled images etc.). If you use the full dataset we recommend to choose different names for the images/results you create yourself to distinguish the old data from the new one you generated yourself.
- RegLib_C02.mrb: input data only, use this to run the tutorial from the start (Slicer mrb file. 19 MB).
- RegLib_C02_full.mrb: includes raw data + all solutions and intermediate files, use to browse/verify (Slicer mrb file. 30 MB).
Keywords
MRI, brain, head, intra-subject, labelmap
Video Screencasts
Procedure
- Compute Registration: open the General Registration (BRAINS) module
- Input Images: Fixed Image Volume: T1
- Input Images: Moving Image Volume: FLAIR
- Output Settings:
- Slicer Linear Transform (create new transform, rename to: "Xf1_Affine")
- Output Image Volume none. No resampling required for linear transforms
- Registration Phases: select/check Rigid , Rigid+Scale, Affine
- Main Parameters:
- increase Number Of Samples to 200,000
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- Align Labelmap: We have now computed the registration transform. To reorient both FLAIR and its label map, we place both volumes inside the transform.
- Place "T1" in the background and "FLAIR" in the foreground. Select "LesionSeg" as the labelmap to display.
- Go to the Data module
- the "FLAIR" image should already be placed within the transform "Xf1_Affine", that was done by the registration module. If not, left click on the "FLAIR" node and drag it onto the "Xf1_Affine" node. You will see a "+" sign appear indicating that the image is placed within the transform.
- Do the same for the "LesionSeg" node, i.e. drag it into the "Xf1_Affine" transform.
- Apply the transforms without resampling / Save result images
- for linear (rigid to affine) transforms you have two options for generating a result image in the new orientation: with and without resampling. The method without resampling leaves the original image data untouched and writes the new orientation into the image header. This preserves the full original image quality and is the preferred method, provided that the software using the result image can read the spatial information in the header correctly. If you open an image with a software that does not account for this header information, the image will appear in its original orientation. The resampling method will create a new image volume by interpolating the data into the new space. This has the advantage that your registered volume will have matching size and resolution as your reference image, but it is associated with some data loss from the interpolation during resampling.
- Apply Transform without resampling:
- Go to the Data module
- left click on the moving image node and drag it into/onto the transform node. You should see a "+" sign appear next to the transform node indicating the volume is now placed under that transform. If the image is in either foreground or background and visible in the slice views or 3D view, you will see the image update to its new position immediately.
- right click on the image node and from the pop-up menu, select Harden Transform. This will apply the transform and move the image node back outside onto the main level.
- double click on the image node and rename it to document that this is no longer the original image but the reoriented one. Note that the image in its original position is replaced in memory and no longer accessible. There is no "undo" for this step, so if you wish to return to the original image, you must reload it from the original file. Be careful not to accidentally overwrite the original image file with the resampled one. Use a new filename when saving the image.
- Alternative: Apply Transform with resampling:
- Go to the BRAINSResample module
- Image To Warp: select FLAIR
- Reference Image: select T1
- Outputs:OutputImage: create & rename new, rename to "FLAIR_Xf1"
- Pixel Type: select short , or match the datatype of the original image.
- Transform File: select "Xf1_Affine"
- Interpolation Mode: select "WindowedSinc" (recommended for 3D MRI). For labelmaps select Nearest Neighbor
- Click Apply. This will generate a new volume with the same resolution as the reference image.
- repeat the same for the label map, but select Nearest Neighbor as the interpolation type.
- Go to the BRAINSResample module
Registration Results
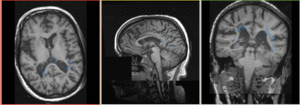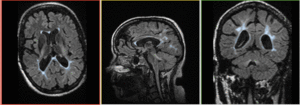 |
baseline & T2 before registration (click to enlarge) |
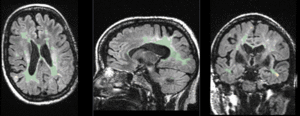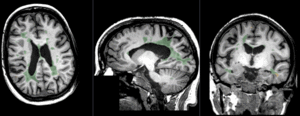 |
baseline to T2 after affine+nonrigid alignment (click to enlarge) |