Difference between revisions of "Documentation:Nightly:Registration:RegistrationLibrary:RegLib C03"
From Slicer Wiki
| Line 14: | Line 14: | ||
|} | |} | ||
| − | + | ==Objective / Background == | |
Goal is to align the DTI image with the structural reference T2 scan that provides accuracte anatomical reference. | Goal is to align the DTI image with the structural reference T2 scan that provides accuracte anatomical reference. | ||
| Line 21: | Line 21: | ||
*[[Documentation/Nightly/Modules/ResampleDTI|''Resample DTI Volume'']] | *[[Documentation/Nightly/Modules/ResampleDTI|''Resample DTI Volume'']] | ||
| − | + | == Alternate Versions == | |
*this example covers the most basic form of directly registering a DTI + baseline to a T2. There is another (more advanced) version that show how to address additional issues of a strong initial rotation and strong voxel-anisotropy for the raw DWI image acquired. [[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C03B|You will find the advanced version here]]. | *this example covers the most basic form of directly registering a DTI + baseline to a T2. There is another (more advanced) version that show how to address additional issues of a strong initial rotation and strong voxel-anisotropy for the raw DWI image acquired. [[Documentation:Nightly:Registration:RegistrationLibrary:RegLib_C03B|You will find the advanced version here]]. | ||
*[http://na-mic.org/Wiki/index.php/Projects:RegistrationLibrary:RegLib_C03 for the Slicer '''4.1''' version of this case see here] | *[http://na-mic.org/Wiki/index.php/Projects:RegistrationLibrary:RegLib_C03 for the Slicer '''4.1''' version of this case see here] | ||
| Line 30: | Line 30: | ||
*[http://slicer.kitware.com/midas3/download/?items=95057,1 '''RegLib_C03.mrb''': includes raw data + all solutions and intermediate files, use to browse/verify <small>(Slicer mrb file. 130 MB). </small>] | *[http://slicer.kitware.com/midas3/download/?items=95057,1 '''RegLib_C03.mrb''': includes raw data + all solutions and intermediate files, use to browse/verify <small>(Slicer mrb file. 130 MB). </small>] | ||
| − | + | == Procedure == | |
| − | This assumes you have the following: 1) a T2 reference image, 2) a DTI baseline image and 3) the DTI volume (both obtained from the [http://www.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/DiffusionTensorEstimation Diffusion Tensor Estimation module]). | + | This assumes you have the following: 1) a T2 reference image, 2) a DTI baseline image and 3) the DTI volume (both obtained from the [http://www.slicer.org/slicerWiki/index.php/Documentation/4.1/Modules/DiffusionTensorEstimation Diffusion Tensor Estimation module]). If you do not have a baseline image, generate a scalar Trace image from the DTI, using the ''Diffusion Tensor Scalar Measurements'' module: |
| − | |||
#open [[DiffusionTensorScalarMeasurements]] | #open [[DiffusionTensorScalarMeasurements]] | ||
#''Input DTI Volume'': select DTI, ''Output Scalar Volume'': create & rename new, rename to "DTI_Trace" or similar | #''Input DTI Volume'': select DTI, ''Output Scalar Volume'': create & rename new, rename to "DTI_Trace" or similar | ||
#''Estimation Parameters'': select ''Trace''; click Apply | #''Estimation Parameters'': select ''Trace''; click Apply | ||
| − | |||
| − | |||
| − | |||
| − | |||
#open [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit Registration : ''General Registration (BRAINS)''] module | #open [http://www.slicer.org/slicerWiki/index.php/Modules:BRAINSFit Registration : ''General Registration (BRAINS)''] module | ||
##''Input Images'': fixed = T2 , moving = DTI_base | ##''Input Images'': fixed = T2 , moving = DTI_base | ||
| Line 49: | Line 44: | ||
##''Registration Phases'': select/check ''Rigid'' , ''Rigid+Scale'', ''Affine'', ''BSpline'' | ##''Registration Phases'': select/check ''Rigid'' , ''Rigid+Scale'', ''Affine'', ''BSpline'' | ||
##''Main Parameters'': | ##''Main Parameters'': | ||
| − | ###increase ''Number Of Samples'' to | + | ###increase ''Number Of Samples'' to 300,000 |
| − | ###set ''B-Spline Grid Size'' to | + | ###set ''B-Spline Grid Size'' to 7,7,5 (we have lower resolution in the IS-direction (z), hence we set a smaller (5) grid size there) |
##Leave all other settings at default | ##Leave all other settings at default | ||
##click: ''Apply''; runtime < 1 min. | ##click: ''Apply''; runtime < 1 min. | ||
Revision as of 15:15, 13 May 2013
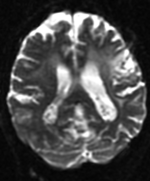
Home < Documentation:Nightly:Registration:RegistrationLibrary:RegLib C03Slicer Registration Library Case #3: Diffusion Weighted Image Volume: align with structural reference MRI
Input

|
|
|

|
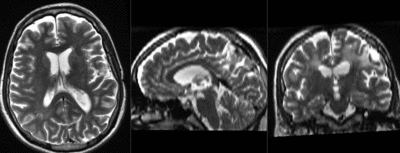
| fixed image 1/target T2 |
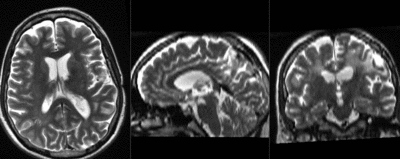
moving image 2a DTI baseline |
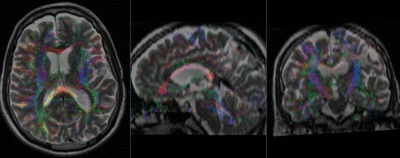
moving image 2b DTI tensor |
Objective / Background
Goal is to align the DTI image with the structural reference T2 scan that provides accuracte anatomical reference.
Modules used
Alternate Versions
- this example covers the most basic form of directly registering a DTI + baseline to a T2. There is another (more advanced) version that show how to address additional issues of a strong initial rotation and strong voxel-anisotropy for the raw DWI image acquired. You will find the advanced version here.
- for the Slicer 4.1 version of this case see here
- for the Slicer 3.6.3 version of this case see here
Download (from NAMIC MIDAS)
- RegLib_C03_raw.mrb: raw data only, use this to run the tutorial from the start (Slicer mrb file. 75 MB).
- RegLib_C03.mrb: includes raw data + all solutions and intermediate files, use to browse/verify (Slicer mrb file. 130 MB).
Procedure
This assumes you have the following: 1) a T2 reference image, 2) a DTI baseline image and 3) the DTI volume (both obtained from the Diffusion Tensor Estimation module). If you do not have a baseline image, generate a scalar Trace image from the DTI, using the Diffusion Tensor Scalar Measurements module:
- open DiffusionTensorScalarMeasurements
- Input DTI Volume: select DTI, Output Scalar Volume: create & rename new, rename to "DTI_Trace" or similar
- Estimation Parameters: select Trace; click Apply
- open Registration : General Registration (BRAINS) module
- Input Images: fixed = T2 , moving = DTI_base
- Output Settings:
- Slicer BSpline Transform (create new transform, rename to: "Xf1_DTbase-T2_BSpline")
- Slicer Linear Transform none
- Output Image Volume (create new volume, rename to: "DTIbaseline_Xf1"
- Registration Phases: select/check Rigid , Rigid+Scale, Affine, BSpline
- Main Parameters:
- increase Number Of Samples to 300,000
- set B-Spline Grid Size to 7,7,5 (we have lower resolution in the IS-direction (z), hence we set a smaller (5) grid size there)
- Leave all other settings at default
- click: Apply; runtime < 1 min.
- Resample DTI
- Open the Resample DTI Volume module (found under: All Modules)
- Input Volume: select DTI
- Output Volume: select create new Diffusion Tensor Volume,and rename it to DTI_Xf1
- Reference Volume: select T2
- Transform Parameters: select transform node "Xf1_DTI-T2_BSpline", for Deformation Field: none ; check the displacement checkbox
- Leave all other settings at defaults
- Click Apply; runtime ~ 2 min.
- set T2 as background and new DTI_Xf1 volume as foreground
- fade between back- and foreground to see DTI overlay onto the T2 image. Note that you can also fade via holding the OPTION+CMD keys (mac) + dragging left mouse.
Registration Results (click to enlarge)
| baseline & T2 before registration | baseline to T2 after affine+nonrigid alignment | DTI and T2 before & after registration |
Keywords
MRI, brain, head, intra-subject, DTI, DWI
Discussion: Key Strategies
- the strong EPI-based distortions of the DTI image make nonrigid registration necessary
- initial alignment & overlap is sufficient so that no "initialization" methods are necessary and registration can succeed without.
- contrast & initial pose are similar enough for registration to succeed without any masking. However the DTI estimation procedure does provide an optional mask that is usually very helpful in registering cases with more "distracting" image content. For an example see the extended version of this case here.
- the DTI in this example is isotropic and hence can be resampled directly. If the DTI contains strong anisotropy of ratios 1:3 or greater, reorienting the DTI can lead to strong artifacts (e.g. in axial direction appear as blue cast in the color orientation view). In that case it is necessary to resample the DWI in the original orientation to an isotropic size before reorienting. It may also be advisable to first reorient the DWI and perform the DTI estimation afterwards.