Difference between revisions of "Documentation/Nightly/Modules/UKFTractography"
(Prepend documentation/versioncheck template. See http://na-mic.org/Mantis/view.php?id=2887) |
m (Text replacement - "https?:\/\/(?:www|wiki)\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+) " to "https://www.slicer.org/wiki/$1 ") |
||
| (19 intermediate revisions by 4 users not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | |||
| − | |||
| − | Contact: | + | {{documentation/{{documentation/version}}/module-acknowledgements}} |
| + | |||
| + | |||
| + | Contact: <email>slicer-users@bwh.harvard.edu</email><br> | ||
| + | Website: https://github.com/pnlbwh/ukftractography<br> | ||
| + | Website: http://slicerdmri.github.io/ | ||
| + | |||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | | | + | |Image:SlicerDMRIScreenshot.jpg|SlicerDMRI |
| − | | | + | |Image:NAC-logo.png|NAC |
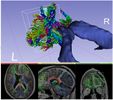
| + | |Image:UKF 1.png|Arcuate fasciculus (AF) tract in the setting of edema [Chen et al, 2015] | ||
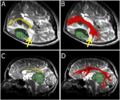
| + | |Image:UKF 2.png|Corpus callosum (CC) tract | ||
}} | }} | ||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-description}} | {{documentation/{{documentation/version}}/module-description}} | ||
| − | |||
| − | |||
| − | |||
| + | For additional references, please see below (References section). | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| + | By default, this module uses a tensor model (either one or two tensors). The default tensor model is a cylinder: both smaller eigenvalues are equal. The NODDI model can also be used. | ||
* 1-Tensor tractography | * 1-Tensor tractography | ||
* 1-Tensor tractography with free water | * 1-Tensor tractography with free water | ||
* 2-Tensor tractography | * 2-Tensor tractography | ||
* 2-Tensor tractography with free water | * 2-Tensor tractography with free water | ||
| + | * Neurite orientation dispersion and density imaging (NODDI) | ||
| Line 60: | Line 50: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | * UKF Tractography tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#UKF <!-- ---------------------------- --> | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-parametersdescription}} | {{documentation/{{documentation/version}}/module-parametersdescription}} | ||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | + | * Tractography Seeding | |
| − | + | * Tractography ROI Seeding | |
| − | + | * Diffusion Tensor Estimation | |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | Reference for 2-tensor tractography: | + | * Reference for 2-tensor tractography: |
| − | * http:// | + | ** Malcolm, J.G., Shenton, M.E. and Rathi, Y., 2010. [http://www.ncbi.nlm.nih.gov/pubmed/20805043 Filtered multitensor tractography. IEEE transactions on medical imaging, 29(9), pp.1664-1675]. |
| − | + | * Reference for 1-tensor and 2-tensor + free-water: | |
| − | Reference for 1-tensor and 2-tensor + free-water: | + | ** Baumgartner C, Michailovich O, Levitt J, Pasternak O, Bouix S, Westin C, Rathi Y. [http://cmic.cs.ucl.ac.uk/cdmri12/pdfs/o1_3.pdf A unified tractography framework for comparing diffusion models on clinical scans. In Computational Diffusion MRI Workshop of MICCAI, Nice 2012 (pp. 27-32).] |
| − | * C | + | * Reference for using UKF in clinical imaging data from tumor patients with edema: |
| − | + | ** Chen Z, Tie Y, Olubiyi O, Rigolo L, Mehrtash A, Norton I, Pasternak O, Rathi Y, Golby AJ, O'Donnell LJ. [http://www.ncbi.nlm.nih.gov/pubmed/26082890 Reconstruction of the arcuate fasciculus for surgical planning in the setting of peritumoral edema using two-tensor unscented Kalman filter tractography. NeuroImage: Clinical. 2015 Dec 31;7:815-22]. | |
| − | + | * Reference for NODDI UKF tractography: | |
| − | + | ** Reddy, C.P. and Rathi, Y., 2016. [http://www.ncbi.nlm.nih.gov/pubmed/27147956 Joint Multi-Fiber NODDI Parameter Estimation and Tractography Using the Unscented Information Filter. Frontiers in Neuroscience, 10]. | |
| − | |||
| Line 209: | Line 78: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| − | + | * https://github.com/pnlbwh/ukftractography | |
| − | + | * https://www.nitrc.org/projects/ukftractography | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| − | {{documentation/{{documentation/version}}/module-footer}} | + | <!-- {{documentation/{{documentation/version}}/module-footer}} --> |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 13:19, 27 November 2019
Home < Documentation < Nightly < Modules < UKFTractography
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Contact: <email>slicer-users@bwh.harvard.edu</email>
| |||||||||
|
Module Description
This module traces fibers in a DWI Volume using the multiple tensor unscented Kalman Filter methodology. At each point on the fiber the most consistent direction is found as a mixture of previous estimates and of the local model.
For more information, please reference: Malcolm, James G., Martha E. Shenton, and Yogesh Rathi. "Filtered multitensor tractography." Medical Imaging, IEEE Transactions on 29.9 (2010): 1664-1675. (http://www.ncbi.nlm.nih.gov/pubmed/20805043)
For additional references, please see below (References section).
Use Cases
By default, this module uses a tensor model (either one or two tensors). The default tensor model is a cylinder: both smaller eigenvalues are equal. The NODDI model can also be used.
- 1-Tensor tractography
- 1-Tensor tractography with free water
- 2-Tensor tractography
- 2-Tensor tractography with free water
- Neurite orientation dispersion and density imaging (NODDI)
Tutorials
- UKF Tractography tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#UKF
Panels and their use
Parameters:
- IO: Input/output parameters
- Input DWI Volume (dwiFile): Input diffusion weighted (DWI) volume
- Input Label Map (seedsFile): Seeds for diffusion. If not specified, full brain tractography will be performed, and the algorithm will start from every voxel in the brain mask where the Generalized Anisotropy is bigger than 0.18
- ROI label to use for seeding (labels): A vector of the ROI labels to be used. There are the voxel values where tractography should be seeded.
- Input brain mask (maskFile): Brain mask for diffusion tractography. Tracking will only be performed inside this mask.
- Output Fiber Bundle (tracts): Output fiber tracts.
- Tractography Options: Basic Parameters
- Seeding: Number of seeds per voxel (seedsPerVoxel): Tractography parameter used in all models. Each seed generates a fiber, thus using more seeds generates more fibers. In general use 1 or 2 seeds, and for a more thorough result use 5 or 10 (depending on your machine this may take up to 2 days to run). Default: 1. Range: 0-50.
- Seeding: Minimum seed FA (seedingThreshold): Tractography parameter used in all models. Seed points whose fractional anisotropy (FA) or mean signal are below this value are excluded. Default: 0.18. Range: 0-1.
- Stopping Criterion: Terminating FA (stoppingFA): Tractography parameter used in tensor model. Tractography will stop when the fractional anisotropy (FA) of the tensor being tracked is less than this value. Note: make sure to also decrease the GA to track through lower anisotropy areas. This parameter is used only in tensor models. Default: 0.15. Range: 0-1.
- Stopping Criterion: Terminating mean signal (stoppingThreshold): Tractography parameter used in all models. Tractography will stop when the mean signal is below this value. Default: 0.1. Range: 0-1.
- Tracking: Number of threads (numThreads): Tractography parameter used in all models. Number of threads used during computation. Set to the number of cores on your workstation for optimal speed. If left undefined, the number of cores detected will be used.
- Tracking: Number of tensors/orientations in model (numTensor): Number of tensors (tensor model) or orientations (NODDI model) used
- Tracking: Step length of tractography (in mm) (stepLength): Tractography parameter used in all models. Step size when conducting tractography. Default: 0.3. Range: 0.1-1.
- Tracking: Rate of change of tensor direction/orientation (Qm): UKF data fitting parameter for tensor or NODDI model: Process noise for angles/direction. Defaults: Noddi-0.001; Single tensor-0.005; other-0.001. Suggested Range: 0.00001 - 0.25. Default of 0.0 indicates the program will assign value based on other model parameters.
- Output: Step length of output tractography (in mm) (recordLength): Tractography parameter used in all models. Step size between points saved along fibers. Default: 0.9. Range: 0.1-4.
- Output: Maximum tract length (in mm) (maxHalfFiberLength): Tractography parameter used in all models. The max length limit of the half fibers generated during tractography. A fiber is "half" when the tractography goes in only one direction from one seed point at a time. Default: 250 mm. Range: 1-500 mm.
- Output: Save Normalized Mean Square Error (recordNMSE): Record output from data fitting: Store normalized mean square error (NMSE) along fibers.
- Tensor Model (default): Tensor model parameters
- Tensor Model: Estimate term for free water (freeWater): Adds a term for free water diffusion to the model. The free water model is a tensor with all 3 eigenvalues equal to the diffusivity of free water (0.003). To output the free water fraction, make sure to use the "save free water" parameter.
- Output: Save tensor FA (recordFA): Record output from tensor model: Save fractional anisotropy (FA) of the tensor(s). Attaches field 'FA' or 'FA1' and 'FA2' for 2-tensor case to fiber.
- Output: Save tensor trace (recordTrace): Record output from tensor model: Save the trace of the tensor(s). Attaches field 'Trace' or 'Trace1' and 'Trace2' for 2-tensor case to fiber.
- Output: Save free water fraction (recordFreeWater): Record output from tensor plus free water model: Save the fraction of free water. Attaches field 'FreeWater' to fiber.
- Output: Save tensors (recordTensors): Record output from tensor model: Save the tensors that were computed during tractography (if using tensor model). The fields will be called 'TensorN', where N is the tensor number. Recording the tensors enables Slicer to color the fiber bundles by FA, orientation, and so on. Recording the tensors also enables quantitative analyses.
- UKF Parameter (Advanced): Rate of change of eigenvalues (Ql): UKF data fitting parameter for tensor model: Process noise for eigenvalues. Defaults: 1 tensor-300 ; 2 tensor-50 ; 3 tensor-100. Suggested Range: 1-1000. Default of 0.0 indicates the program will assign value based on other model parameters.
- UKF Parameter (Advanced): Rate of change of freewater weight (Qw): UKF data fitting parameter for tensor plus free water model: Process noise for free water weights, ignored if no free water estimation. Defaults: 1 tensor-0.0025; 2 tensor-0.0015. Suggested Range: 0.00001-0.25. Default of 0.0 indicates the program will assign value based on other model parameters.
- NODDI Model: Use NODDI model
- NODDI Model: Use NODDI Model (noddi): Use neurite orientation dispersion and density imaging (NODDI) model instead of tensor model.
- Output: Save NODDI intra-cellular volume fraction. (recordVic): Record output from NODDI model: Store volume fraction of intra-cellular compartment along fibers.
- Output: Save NODDI dispersion parameter (kappa) (recordKappa): Record output from NODDI model: concentration parameter that measures the extent of orientation dispersion.
- Output: Save NODDI CSF volume fraction. (recordViso): Record output from NODDI model: Store volume fraction of CSF compartment along fibers.
- UKF Parameter (Advanced): Rate of change of kappa value (Qkappa): UKF data fitting parameter for NODDI model: Rate of change of kappa (orientation dispersion) value. Higher kappa values indicate more fiber dispersion. Default: 0.01.
- UKF Parameter (Advanced): Rate of change of intracellular volume fraction (Qvic): UKF data fitting parameter for NODDI model: Rate of change of volume fraction of intracellular component. Default: 0.004.
- Signal Parameters (Expert Only):
- Signal Parameter (Advanced): Expected noise in signal (Rs): UKF Data Term: Measures expected noise in signal. This is used by the UKF method to decide how much to trust the data. This should be increased for very noisy data or reduced for high quality data. Defaults: single tensor/orientation-0.01; other-0.02. Suggested Range: 0.001-0.25. Default of 0.0 indicates the program will assign value based on other model parameters.
- Not Used: Debug/Develop Only :
- Signal Parameter (Advanced): Sigma for Gaussian interpolation of signal (sigmaSignal): UKF Data Term: Sigma for Gaussian kernel used to interpolate the signal at sub-voxel locations. Default: 0.0
- Record states (recordState): Develop/Debug Only: Store the states along the fiber. Will generate field 'state'. The state is the model for UKF. In the case of the two tensor model, it is a ten-parameter vector.
- Record the covariance matrix (recordCovariance): Develop/Debug Only: Store the covariance matrix along the fiber. Will generate field 'covariance' in fiber. This is the covariance from the unscented Kalman filter.
- Use full tensor model (fullTensorModel): Develop/Debug Only: Use the full tensor model instead of the default model. The default model has both smaller eigenvalues equal, whereas the full model allows 3 different eigenvalues.
- Maximum branching angle (maxBranchingAngle): Develop/Debug Only: Maximum branching angle, in degrees. When using multiple tensors, a new branch will be created when the tensors' major directions form an angle between (minBranchingAngle, maxBranchingAngle). Branching is supressed when this maxBranchingAngle is set to 0.0. Default: 0.0. Range: 0-90.
- Minimum branching angle (minBranchingAngle): Develop/Debug Only: Minimum branching angle, in degrees. When using multiple tensors, a new branch will be created when the tensors' major directions form an angle between (minBranchingAngle, maxBranchingAngle). Default: 0. Range: 0-90.
- Branched Fibers (second tensor, optional) (tractsWithSecondTensor): Develop/Debug Only: Tracts generated, with second tensor output (if there is one)
- Store tensors' main directions (storeGlyphs): Develop/Debug Only: Store tensors' main directions as two-point lines in a separate file named glyphs_{tracts}. When using multiple tensors, only the major tensors' main directions are stored
- Write Binary Tracts File (writeAsciiTracts): Develop/Debug Only: Write tract file as ASCII text format. Default is not ASCII. Default tracts are written on VTK binary data file.
- Write uncompressed Tracts File (writeUncompressedTracts): Develop/Debug Only: Write tract file as a VTK uncompressed data file
- DEPRECATED REMOVED: minGA (was Stopping Criterion: Terminating GA):
DEPRECATED REMOVED: this parameter is no longer valid! Please use 'stoppingThreshold' instead! GA is no longer used as a stopping threshold. Please see https://github.com/pnlbwh/ukftractography/pull/64 for more information. (Was: Tractography parameter used in all models. Tractography will stop when the generalized anisotropy (GA) is less than this value. GA is a normalized variance of the input signals (so it does not depend on any model). Note: to extend tracking through low anisotropy areas, this parameter is often more effective than the minFA. This parameter is used by both tensor and NODDI models. Default: 0.1. Range: 0-1.)
- Allow in-memory data transfer (AllowMemoryTransfer): Allow in-memory data transfer
List of parameters generated transforming this XML file using this XSL file. To update the URL of the XML file, edit this page.
Similar Modules
- Tractography Seeding
- Tractography ROI Seeding
- Diffusion Tensor Estimation
References
- Reference for 2-tensor tractography:
- Malcolm, J.G., Shenton, M.E. and Rathi, Y., 2010. Filtered multitensor tractography. IEEE transactions on medical imaging, 29(9), pp.1664-1675.
- Reference for 1-tensor and 2-tensor + free-water:
- Baumgartner C, Michailovich O, Levitt J, Pasternak O, Bouix S, Westin C, Rathi Y. A unified tractography framework for comparing diffusion models on clinical scans. In Computational Diffusion MRI Workshop of MICCAI, Nice 2012 (pp. 27-32).
- Reference for using UKF in clinical imaging data from tumor patients with edema:
- Chen Z, Tie Y, Olubiyi O, Rigolo L, Mehrtash A, Norton I, Pasternak O, Rathi Y, Golby AJ, O'Donnell LJ. Reconstruction of the arcuate fasciculus for surgical planning in the setting of peritumoral edema using two-tensor unscented Kalman filter tractography. NeuroImage: Clinical. 2015 Dec 31;7:815-22.
- Reference for NODDI UKF tractography:
- Reddy, C.P. and Rathi, Y., 2016. Joint Multi-Fiber NODDI Parameter Estimation and Tractography Using the Unscented Information Filter. Frontiers in Neuroscience, 10.
Information for Developers