Difference between revisions of "Documentation/Nightly/Modules/TractographyInteractiveSeeding"
(Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
m (Text replacement - "\bhttps:\/\/www\.slicer\.org\/slicerWiki\/index\.php\/([^ ]+)\b " to "https://www.slicer.org/wiki/$1 ") |
||
| (7 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | + | ||
| − | + | ||
| − | + | Title: Tractography Interactive Seeding | |
| − | + | ||
| − | Contact: | + | Author(s)/Contributor(s): Alex Yarmarkovich, Steve Pieper, Demian Wassermann, Isaiah Norton (SPL, LMI, BWH, SlicerDMRI) |
| + | |||
| + | License: 3D Slicer Contribution and Software License Agreement | ||
| + | |||
| + | Acknowledgements: The SlicerDMRI developers gratefully acknowledge funding for this project provided by NIH NCI ITCR U01CA199459 (Open Source Diffusion MRI Technology For Brain Cancer Research), NIH P41EB015898 (National Center for Image-Guided Therapy) and NIH P41EB015902 (Neuroimaging Analysis Center), as well as the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. | ||
| + | |||
| + | Contact: slicer-users@bwh.harvard.edu | ||
| + | |||
| + | Website: http://slicerdmri.github.io/ | ||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| + | |Image:SlicerDMRIScreenshot.jpg|SlicerDMRI | ||
|Image:Logo-splnew.jpg|Surgical Planning Laboratory | |Image:Logo-splnew.jpg|Surgical Planning Laboratory | ||
| + | |Image:NAC-logo.png|NAC | ||
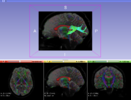
| + | |Image:Fiducials seeding 1.png| Tractography Fiducial Seeding | ||
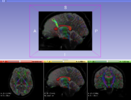
| + | |Image:Fiducials seeding 2.png| Tractography Fiducial Seeding | ||
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 22: | Line 35: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module is used for interactive seeding of DTI fiber tracts starting from a list of fiducial points or vertices of a model, or a label map volume | + | This module is used for interactive seeding of DTI fiber tracts starting from a list of fiducial points or vertices of a model, or a label map volume. |
| Line 35: | Line 48: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | Links to tutorials that use this module: | |
| − | + | * Slicer4 Diffusion Tensor Imaging Tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial * Slicer4 Neurosurgical Planning Tutorial: https://www.slicer.org/wiki/Documentation/4.5/Training#Slicer4_Neurosurgical_Planning_Tutorial <!-- ---------------------------- --> | |
| − | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | |||
| − | |||
{|style="width: 100%" | {|style="width: 100%" | ||
| Line 52: | Line 62: | ||
** Output FiberBundle: Select the bundle that you want to create or modify from a list. | ** Output FiberBundle: Select the bundle that you want to create or modify from a list. | ||
** Enable Seeding Tracts: A toggle to enable/disable seeding. Disable if you want to change multiple parameters without re-running module at each step. | ** Enable Seeding Tracts: A toggle to enable/disable seeding. Disable if you want to change multiple parameters without re-running module at each step. | ||
| − | * | + | * Seed Placement Option panel: |
| − | ** | + | ** Fiducial Region Size: The size of the box around each fiducial used for seeding (mm). |
| − | ** | + | ** Fiducial Seeding Step Size: Step between the seeding samples in the box (mm). |
** Seed Selected Fiducials: Flag indicating whether to use only selected fiducials for seeding or all of them. | ** Seed Selected Fiducials: Flag indicating whether to use only selected fiducials for seeding or all of them. | ||
** Maximum Number of Seeds: Used with models to specify the limit of the number of vertices to be used for seeding. | ** Maximum Number of Seeds: Used with models to specify the limit of the number of vertices to be used for seeding. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
* Tractography Seeding Parameters panel: | * Tractography Seeding Parameters panel: | ||
** Minimum Path Length: Select only tracts that are longer than this value (mm). | ** Minimum Path Length: Select only tracts that are longer than this value (mm). | ||
| Line 87: | Line 80: | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
* Tractography Label Map Seeding | * Tractography Label Map Seeding | ||
| + | * UKF Tractography | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | + | * Interactive Diffusion Tensor Tractography Visualization for Neurosurgical Planning: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3112275/ | |
| − | + | * http://slicerdmri.github.io/ | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| − | + | * https://github.com/SlicerDMRI | |
Latest revision as of 14:01, 27 November 2019
Home < Documentation < Nightly < Modules < TractographyInteractiveSeeding
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Author(s)/Contributor(s): Alex Yarmarkovich, Steve Pieper, Demian Wassermann, Isaiah Norton (SPL, LMI, BWH, SlicerDMRI) License: 3D Slicer Contribution and Software License Agreement Acknowledgements: The SlicerDMRI developers gratefully acknowledge funding for this project provided by NIH NCI ITCR U01CA199459 (Open Source Diffusion MRI Technology For Brain Cancer Research), NIH P41EB015898 (National Center for Image-Guided Therapy) and NIH P41EB015902 (Neuroimaging Analysis Center), as well as the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Contact: slicer-users@bwh.harvard.edu Website: http://slicerdmri.github.io/ | |||||||||||
|
Module Description
This module is used for interactive seeding of DTI fiber tracts starting from a list of fiducial points or vertices of a model, or a label map volume.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Create DTI fiber tracts starting from a list of fiducial points. Tracks created from a small number of fiducial points are are highly interactive and can be used in exploration mode while moving fiducials with the help of transformations. This mode can be useful in pre-op planning or for tracking tools during the surgery.
- Use Case 2: Create DTI fiber tracts starting from vertices of a model. This mode can be useful for exploring tracts around a tumor model.
- Use Case 2: Create DTI fiber tracts starting from a label map volume. Tracks created from a label map ROI can be used in interactive mode while modifying the label map using Editor module.
Tutorials
Links to tutorials that use this module:
- Slicer4 Diffusion Tensor Imaging Tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#Slicer4_Diffusion_Tensor_Imaging_Tutorial * Slicer4 Neurosurgical Planning Tutorial: https://www.slicer.org/wiki/Documentation/4.5/Training#Slicer4_Neurosurgical_Planning_Tutorial
Panels and their use
Similar Modules
References
Information for Developers
|