Difference between revisions of "Documentation/Nightly/Modules/T1Mapping"
Stevedaxiao (talk | contribs) |
Stevedaxiao (talk | contribs) |
||
| Line 18: | Line 18: | ||
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, and by National Cancer Institute as part of the Quantitative Imaging Network initiative (U01CA154601) and QIICR (U24CA180918).<br> | This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, and by National Cancer Institute as part of the Quantitative Imaging Network initiative (U01CA154601) and QIICR (U24CA180918).<br> | ||
| − | Implementation of the T1 Mapping was contributed by Xiao Da from MGH | + | Implementation of the T1 Mapping was contributed by Xiao Da from MGH. <br> |
Author: Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH) and Jayashree Kalpathy-Cramer (MGH)<br> | Author: Xiao Da (MGH), Yangming Ou (MGH), Andriy Fedorov (BWH) and Jayashree Kalpathy-Cramer (MGH)<br> | ||
Revision as of 03:47, 11 September 2015
Home < Documentation < Nightly < Modules < T1Mapping
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
| |||||||
|
Module Description
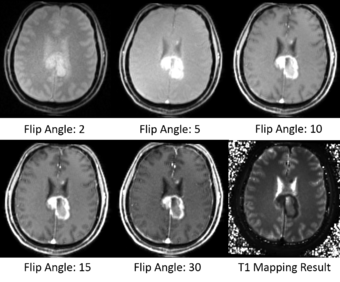
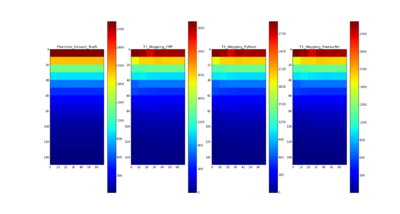
T1 mapping estimates effective tissue parameter maps (T1) from multi-spectral FLASH MRI scans with different flip angles. T1 mapping can be used to optimize parameters for a sequence, monitor diseased tissue, measure Ktrans in DCE-MRI and etc.
The method and equations for Variable Flip Angle T1 Mapping are available here: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3620726/pdf/nihms-423474.pdf
Use Cases
- Take multi-spectral FLASH images with an arbitrary number of flip angles as input, and estimate the T1 values of the data for each voxel.
- Read repetition time(TR), echo time(TE) and flip angles from the Dicom header directly.
- Prostate, brain, head & neck, cervix, breast and etc.
Tutorials
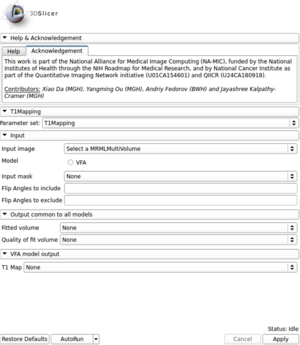
Panels and their use
|
Similar Modules
References
Information for Developers
| Section under construction. |
Source code: https://github.com/stevedaxiao/T1_Mapping_CPP