Difference between revisions of "Documentation/Nightly/Modules/SlicerToKiwiExporter"
m (Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
|||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Introduction and Acknowledgements}} | ||
| + | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
| + | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| + | '''Extension:''' [[Documentation/{{documentation/version}}/Extensions/SlicerToKiwiExporter|SlicerToKiwiExporter]]<br> | ||
| + | '''Acknowledgments:''' | ||
| + | This work is part of the National Alliance for Medical Image Computing (NAMIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149.<br> | ||
| + | '''Author:''' Jean-Christophe Fillion-Robin ({{collaborator|name|kitware}})<br> | ||
| + | '''Contributor1:''' Pat Marion ({{collaborator|name|kitware}})<br> | ||
| + | '''Contributor2:''' Steve Pieper ({{collaborator|name|isomics}})<br> | ||
| + | '''Contributor3:''' Atsushi Yamada (Shiga University of Medical Science)<br> | ||
| + | '''Contact:''' Jean-Christophe Fillion-Robin, <email>jchris.fillionr@kitware.com</email><br> | ||
| + | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |||
|{{collaborator|logo|kitware}}|{{collaborator|longname|kitware}} | |{{collaborator|logo|kitware}}|{{collaborator|longname|kitware}} | ||
| − | |{{collaborator|logo|namic}}|{{collaborator|longname| | + | |{{collaborator|logo|namic}}|{{collaborator|longname|isomics}} |
| + | |Image:SumsLogo2014.jpg|Shiga University of Medical Science | ||
}} | }} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 28: | Line 28: | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| + | The SlicerToKiwiExporter module provides Slicer user with any easy way to export [[Documentation/{{documentation/version}}/Modules/Models|models]] into a [http://www.kiwiviewer.org/ KiwiViewer] scene file. | ||
| + | |||
| + | [http://www.kiwiviewer.org/ KiwiViewer] is a free, open-source visualization app for exploring scientific and medical datasets that runs on Android and iOS mobile devices with multi-touch interaction. [http://www.kiwiviewer.org/ KiwiViewer] supports a variety of file formats, including obj, stl, ply, and vtk. Datasets may be loaded into [http://www.kiwiviewer.org/ KiwiViewer] from the SD card, email attachments, or Midas, or DropBox, or downloaded from a URL. | ||
| + | The following tutorials will provide steo-by-step instructions to: | ||
| + | * [[#Saving_a_MRML_scene_as_KiwiViewer_scene_file|Export a Slicer MRML scene into KiwiViewer scene file]] | ||
| + | * [[#Uploading_KiwiViewer_Scene_file_to_Midas|Upload the exported file onto a shared data source]] | ||
| + | * [[#Loading_KiwiViewer_Scene_file_from_Midas|Load the exported file into KiwiViewer]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | + | ||
| − | + | Interactively show an audience models created using slicer on an iPad. | |
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| + | |||
| + | == Saving a MRML scene as KiwiViewer scene file == | ||
| + | <gallery widths=450px perrow=2> | ||
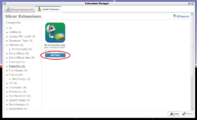
| + | File:SlicerToKiwiExporter-Slicer-install.png|Install extension named "SlicerToKiwiExporter". [[Documentation/{{documentation/version}}/SlicerApplication/ExtensionsManager#Installing_an_extension|Need help?]] | ||
| + | File:SlicerToKiwiExporter SaveDialog Select-file-format 1.png| (1) Open save dialog and (2) select <code>KiwiViewer File (*.kiwi.zip)</code> file format. | ||
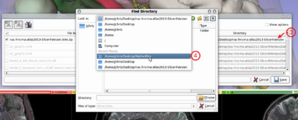
| + | File:SlicerToKiwiExporter SaveDialog Select-dest-directory 2.png| (3 & 4) Choose destination directory | ||
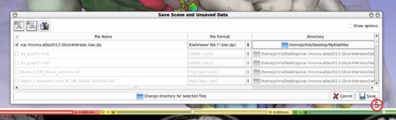
| + | File:SlicerToKiwiExporter SaveDialog ClickSave 3.png| (5) Click on "Save" to export the MRML scene. | ||
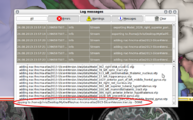
| + | File:SlicerToKiwiExporter_CheckLog_4.png| Open error dialog (View -> Error Log) and check log. | ||
| + | In this example, we can confirm that the MRML scene has been exported to <code>/home/jchris/Desktop/MyKiwiFiles/nac-hncma-atlas2013-Slicer4Version.kiwi.zip</code> | ||
| + | </gallery> | ||
| + | |||
| + | == Uploading KiwiViewer Scene file to Midas == | ||
| + | |||
| + | <ol start="1" style="list-style-type: decimal;"> | ||
| + | |||
| + | {{:Documentation/{{documentation/version}}/Developers/Tutorials/CreateMidasAccount/steps}} | ||
| + | |||
| + | <li><p>If not already done, send an email on the [http://massmail.bwh.harvard.edu/mailman/listinfo/slicer-user slicer users list] asking to be granted write permission on the [http://slicer.kitware.com/midas3/folder/1682 KiwiViewer folder].</p> | ||
| + | <pre> | ||
| + | To: slicer-user@bwh.harvard.edu | ||
| + | Subject: Asking permission to write to the KiwiViewer folder | ||
| + | Hi, | ||
| + | |||
| + | I am user KiwiView application to [...] | ||
| + | |||
| + | Could you grant user YourUserName write access to the KiwiViewer folder ? | ||
| + | |||
| + | </pre></li> | ||
| + | |||
| + | <li><p>Go to http://slicer.kitware.com, click on Communities (1) and select "NA-MIC" (2)</p> | ||
| + | [[File:SlicerToKiwiExporter-UploadMidas SelectNAMIC Community.png|450px]] | ||
| + | |||
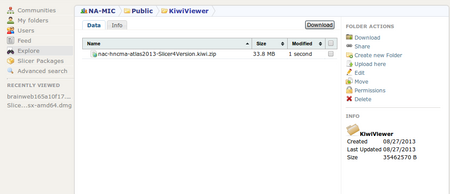
| + | <li><p>Browse, select the destination folder "Kiwiviewer" (3) and click on "Upload here" (4)</p> | ||
| + | [[File:SlicerToKiwiExporter-UploadMidas Upload.png|450px]] | ||
| + | </li> | ||
| + | |||
| + | <li><p>Select one or more files (5) and click "Upload" button (6)</p> | ||
| + | [[File:SlicerToKiwiExporter-UploadMidas SelectFile-and-upload.png|450px]] | ||
| + | </li> | ||
| + | |||
| + | <li><p>Upload is complete</p> | ||
| + | [[File:SlicerToKiwiExporter-UploadMidas complete.png|450px]] | ||
| + | </li> | ||
| + | |||
| + | </ol> | ||
| + | |||
| + | == Loading KiwiViewer Scene file from Midas == | ||
| + | |||
| + | <gallery widths=265px heights=350px perrow=4> | ||
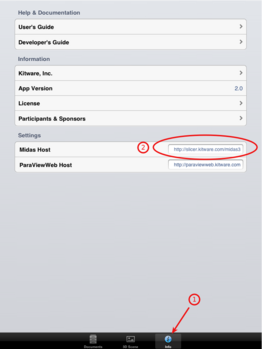
| + | File:SlicerToKiwiExporter Kiwiviewer 1.PNG|Tap on "Info" (1) and enter "http://slicer.kitware.com/midas3" for "Midas Host" | ||
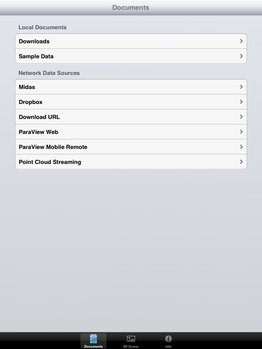
| + | File:SlicerToKiwiExporter Kiwiviewer 2.PNG|Tap on "Documents", then tap on "Midas" in the available Network Data Sources | ||
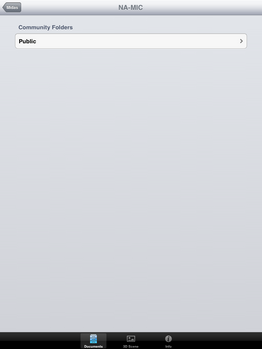
| + | File:SlicerToKiwiExporter Kiwiviewer 3.PNG|Tap on "NA-MIC" | ||
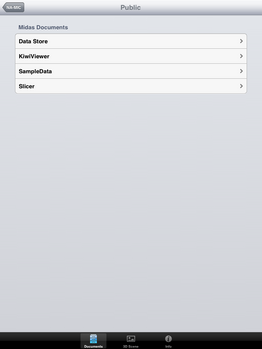
| + | File:SlicerToKiwiExporter Kiwiviewer 4.PNG|Tap on "Public" | ||
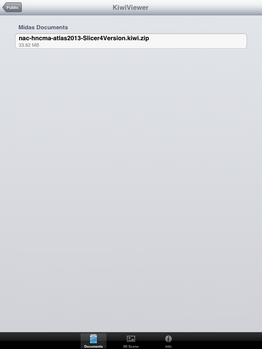
| + | File:SlicerToKiwiExporter Kiwiviewer 5.PNG|Tap on "KiwiViewer" | ||
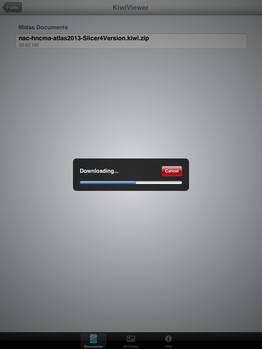
| + | File:SlicerToKiwiExporter Kiwiviewer 6.PNG|Tap on KiwiViewer scene file to download and visualize. | ||
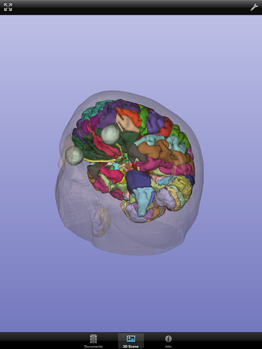
| + | File:SlicerToKiwiExporter Kiwiviewer 7.PNG|Download in progress ... | ||
| + | File:SlicerToKiwiExporter Kiwiviewer 8.PNG|Downloaded file is automatically loaded | ||
| + | File:SlicerToKiwiExporter Kiwiviewer 9.PNG|Tap on "Settings" icon to see data properties | ||
| + | </gallery> | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 54: | Line 112: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| + | |||
| + | Source: https://github.com/jcfr/SlicerToKiwiExporter | ||
| + | |||
{{documentation/{{documentation/version}}/module-developerinfo}} | {{documentation/{{documentation/version}}/module-developerinfo}} | ||
Latest revision as of 00:57, 2 April 2015
Home < Documentation < Nightly < Modules < SlicerToKiwiExporter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: SlicerToKiwiExporter | |||||||
|
Module Description
The SlicerToKiwiExporter module provides Slicer user with any easy way to export models into a KiwiViewer scene file.
KiwiViewer is a free, open-source visualization app for exploring scientific and medical datasets that runs on Android and iOS mobile devices with multi-touch interaction. KiwiViewer supports a variety of file formats, including obj, stl, ply, and vtk. Datasets may be loaded into KiwiViewer from the SD card, email attachments, or Midas, or DropBox, or downloaded from a URL.
The following tutorials will provide steo-by-step instructions to:
- Export a Slicer MRML scene into KiwiViewer scene file
- Upload the exported file onto a shared data source
- Load the exported file into KiwiViewer
Use Cases
Interactively show an audience models created using slicer on an iPad.
Tutorials
Saving a MRML scene as KiwiViewer scene file
Install extension named "SlicerToKiwiExporter". Need help?
Uploading KiwiViewer Scene file to Midas
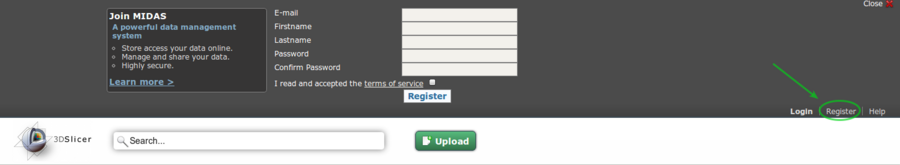
Create an account on the extension server: http://slicer.kitware.com by clicking on the
Registerlink in the top right cornerGo to NA-MIC community and click on
Join communityIf not already done, send an email on the slicer users list asking to be granted write permission on the KiwiViewer folder.
To: slicer-user@bwh.harvard.edu Subject: Asking permission to write to the KiwiViewer folder Hi, I am user KiwiView application to [...] Could you grant user YourUserName write access to the KiwiViewer folder ?
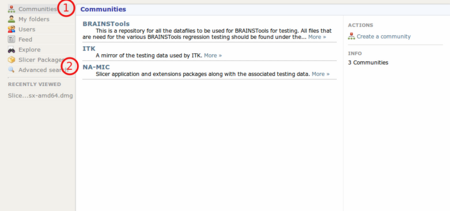
Go to http://slicer.kitware.com, click on Communities (1) and select "NA-MIC" (2)
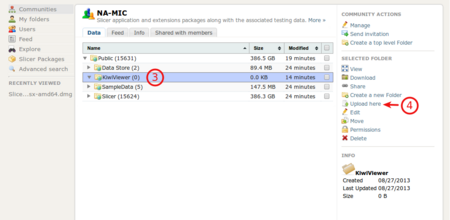
Browse, select the destination folder "Kiwiviewer" (3) and click on "Upload here" (4)
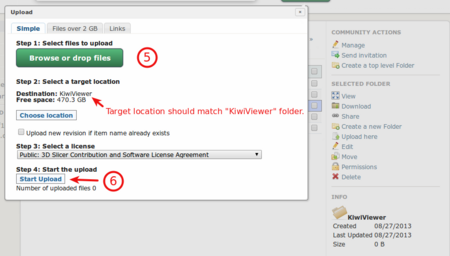
Select one or more files (5) and click "Upload" button (6)
Upload is complete
This image shows the top portion of http://slicer.kitware.com after it has been expanded by clicking the Register button.
Loading KiwiViewer Scene file from Midas
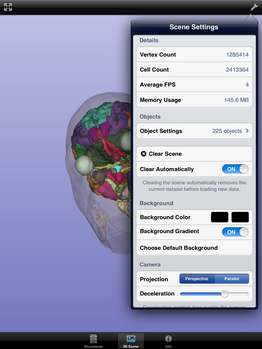
Tap on "Info" (1) and enter "http://slicer.kitware.com/midas3" for "Midas Host"
Panels and their use
Similar Modules
References
Information for Developers
Source: https://github.com/jcfr/SlicerToKiwiExporter
| Section under construction. |