Home < Documentation < Nightly < Modules < ShapeVariationAnalyzer
Introduction and Acknowledgements
|
Extension: ShapeVariationAnalyzer
Acknowledgments: This work was supported by the National Institutes of Dental and Craniofacial Research and Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number R01DE024450.
Author: Priscille de Dumast (University of Michigan)
Contributor1: Laura Pascal (University of Michigan)
Contributor2: Lucia Cevidanes (University of Michigan)
Contributor3: Juan Carlos Prieto (University of North Carolina)
Contact: Priscille de Dumast, <email>prisgdd@umich.edu</email>
License: Apache License, Version 2.0
|
|
|
Module Description
ShapeVariationAnalyzer allows the classification of 3D models, according to their morphological variation. This classifier can be used to identify shapes, help to provide a diagnosis, evaluate the staging of a disease.
This tool is based on a deep learning neural network which evaluates all the possible options and learns from past experience. The aim of this extension is to help understand morphological variations and provide an idea of the classification.
In a first phase, the neural network needs to learn from well-known shapes, in order to be able to process new models in the classification phase.
The module is composed of multiple panels to perform the different steps of the process: create the classification groups, compute their average shapes, train the classifier and classify shapes.
What is an artificial neural network?
A neural network is a computing system, inspired by our own human brain. It learns from a large dataset (training dataset) containing both the input and the expected output (in our case, the 3D model and its morphology classification). During the training, the network’s settings are adjusted until we achieve automatic classification that matches the expert classification.
The network can then be tested with any input, preferably not from the training dataset. This allows to evaluate the performance of the network.
Use Cases
 Logo for ShapeVariationAnalyzer |
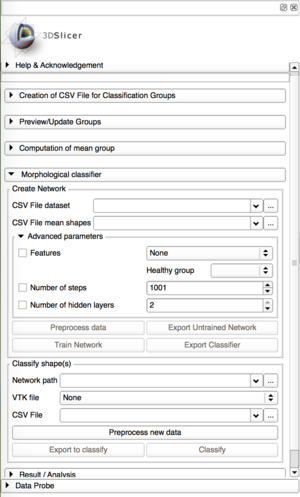 Interface of ShapeVariationAnalyzer |
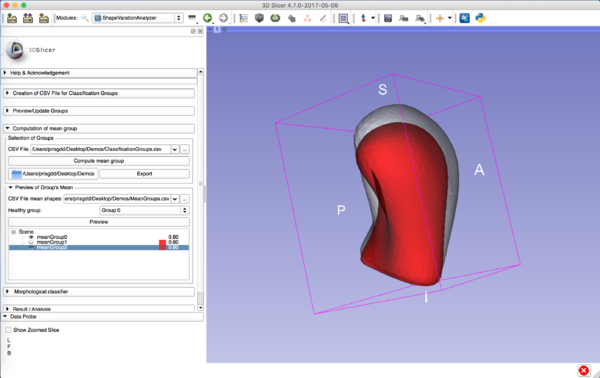 ShapeVariationAnalyzer in Slicer |
Tutorials
Requirements
To use this extension on Windows, you must run 3D Slicer as administrator.
Prerequisities
The classification can be performed with as many groups as desired.
Please note that the amount of data is very important during the training phase. The more data you have, the more accurate the classification will be. Moreover, it is better to have a dataset equally divided into the classes.
Both the 3D models into the training dataset and those to classify must have the same number of points. The most correspondent the shapes are, the better it is for the computation.
This can be performed thanks to ShapeAnalysisModule and improved with RigidAlignment/Groups.
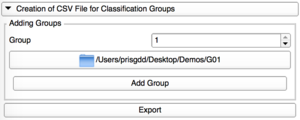
Creation of CSV File

|
This tab allows the user to create a CSV file gathering all the VTK file paths and their corresponding group.
This CSV file lists all the files used in the training dataset. It will be necessary at each step of the classification.
To create this file, we recommend that
The file is automatically loaded into the next steps when it is exported.
|
|
|
|
|
Add Group
Add one by one the directories containing the VTK files for each group.
For a new group, the group number is automatically selected.
|
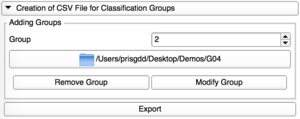
Modify Group
Each group can be edited, with a modification of its corresponding repository.
For this, review the group already added thanks to the spin box.
|
Remove Group
Only the last group added can be removed.
Select the group with the spinbox and remove it.
|
Preview/Update of the classification groups
|
This tab allows the visualization of the 3D models in ShapePopulationViewer, and update the group assigned to the shapes if needed.
If the groups are updated, a new CSV file will be generated.
|
- Preview with ShapePopulationViewer
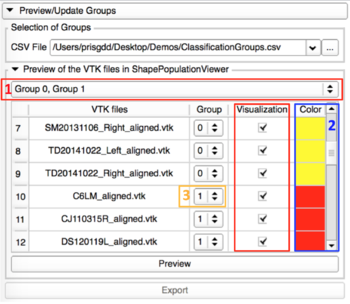
|
- Select the groups or specific shapes you would like to visualize
- Colors that the meshes will have in Shape Population Viewer with the attribute "DisplayClassificationGroup"
- Possibility to change the group of a shape thanks to the spinbox in the group row.
- Clic on the Preview button, and select all the VTK Files in the pop-up window
- If some changes were made on the groups, please export those modifications.
|
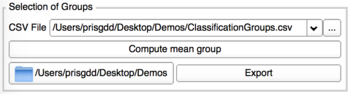
Computation of mean groups
|
This tab allows the computation of the average shape of each group, export those resulting models locally and allows their visualization into 3D Slicer.
You’ll need to provide the CSV file which contains the classification groups.
The exported CSV file will be automatically loaded into the next step.
|
|
- Load the CSV file containing the classification groups, if you haven't performed the previous steps
- Clic on Compute mean group
- Chose a repository to export the average shapes and a CSV File listing them.
Average shapes : meanGroup[i].vtk
CSV File mean shapes : MeanGroups.csv
|
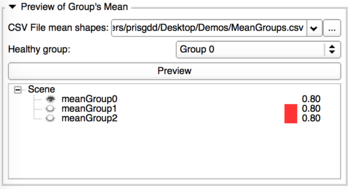
|
- Load the CSV file containing all the average shapes if you haven't performed the computation step.
- Select the control group - it will appear in a different color.
- Clic on Preview to visualize the average shapes
- Check the eye icon on the left to choose which shape you want to display
|
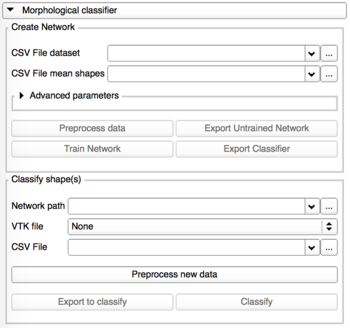
Morphological classification
|
This tab is divided into 2 panels:
- The creation and the training of the classifier
- The classification using a neural network previously trained
|
|
Preprocessing the data
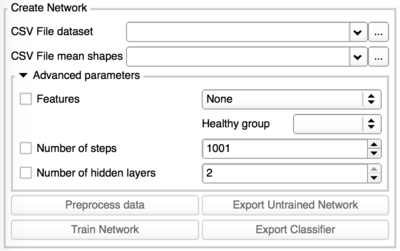
The neural network needs to know from which specific features of the shapes it should learn.
Those features are extracted from the shape during that preprocessing phase.
In the Advanced parameters box, the user can select the features he thinks are the most relevant. Only those selected will be used during the training and in future classification based on this classifier. If the Advanced parameters tab is closed, by default all the features are used.
The possible features are: Curvedness, Normals, Maximum Curvature, Minimum Curvature, Mean Curvature, Gaussian Curvature, Shape Index, Distances to average shapes.
Training the network
The training time of the classifier depends on the number of shapes, their size, the number of groups and the advanced parameters. This can take a while. Be patient! ;)
At the end of the training, the estimated accuracy is displayed. If you are satisfied, you can export the Classifier, if not, you can try to re-train the network using different parameters or features.
When you click on Export Network, this will create a zip file in the chosen directory, containing everything necessary for later classification.
Please do not modify it, at risk that it no longer functions afterward.
TODO: Choice of more advanced network parameters
|
|
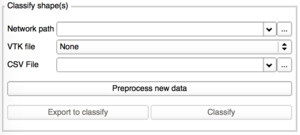
|
To classify one or more shapes, you would need:
- The network previously trained (zipfile)
- The VTK file of the shape you would like to classify OR a CSV file containing all the paths to the VTK you want to classify.
Please note that classifying too many shapes at the same time (via CSV file), can stop Slicer.
At the end of the classification, the results are automatically displayed in the next tab Results/Analysis.
|
Results/Analysis
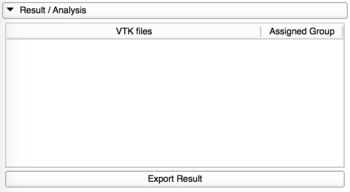
|
Results of the classification are automatically loaded into this tab when the computation is done.
|
Similar Modules
N/A
References
N/A
Information for Developers
The source code of this module is available on GitHub: [1]
The source code of the CLI used to compute an average shape and extract the features is available on Github, independently of the module: [2]