Difference between revisions of "Documentation/Nightly/Modules/SegmentationAidedRegistration"
(panel and parameters) |
(tutorial) |
||
| Line 55: | Line 55: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | * Step 1. Give the input images, including: | |
| + | ** Fixed grayscale image | ||
| + | ** Segmentation label image for the target region in the fixed image | ||
| + | ** Moving grayscale image | ||
| + | ** Segmentation label image for the target region in the moving image | ||
| + | * Step 2. Give the output filename. | ||
| + | * Run | ||
| + | * There is an optional parameter: | ||
| + | ** Only perform deformable registration locally? Yes by default. The output registered grayscale image will only be around the target region. Uncheck to get the registered moving image in the entire region, but this is VERY SLOW. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 21:03, 18 June 2013
Home < Documentation < Nightly < Modules < SegmentationAidedRegistration
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. Contributor: Yi Gao, Brigham and Women's Hospital | |||||||||
|
Module Description
When we want to register two images, often times, we would like an accurate matching at certain region. For example, in the atrial fibrillation longitudinal study, we want to register the pre-op and post-op images. In particular, we want the matching at the atrium to be emphasized. In such cases, we can use the segmentation of the target region, atrium in the previous example, to aid the registration process. This module is such a segmentation aided registration tool.
Use Cases
- In the atrial fibrillation longitudinal study, we want to register the pre-op and post-op images. In particular, we want the matching at the atrium to be emphasized.
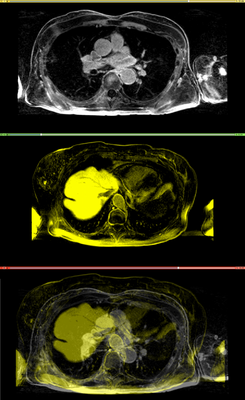
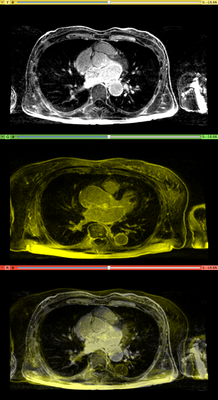
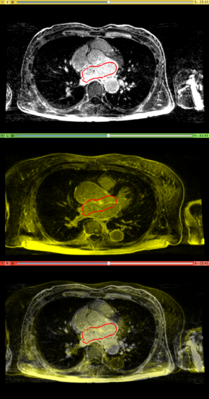
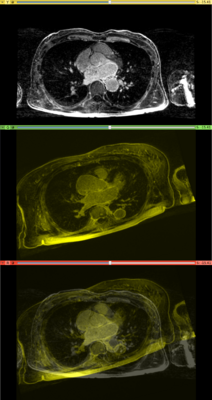
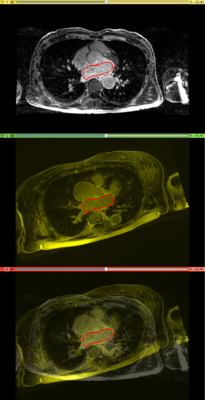
Affine registration using MI metric of the two LGE-MR images, with the red contour showing the left atrium. Top: fixed image; Middle: registered moving image; Bottom: putting them together. We see that the overall registration is good, but the left atrium position is not accurate. In the case where we want to study the change of LA at those two time points, foucs has been put on the LA region.
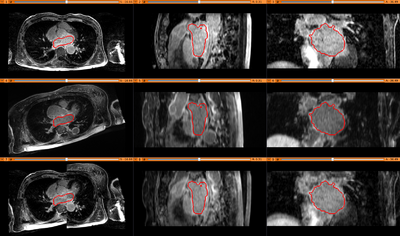
Results. Top row: Pre-op LGE-MRI with its segmentation of the left atrium (LA). Middle row: Pre-op LA contour overlayed on the registered Post-op LGE-MRI. See the matching of the LA region. Bottom row: checker-board view of Pre-op and registered Post-op LGE-MRIs: they match well at the LA region but badly otherwise, indicating the necessity of using segmentation to aid the registration.
Tutorials
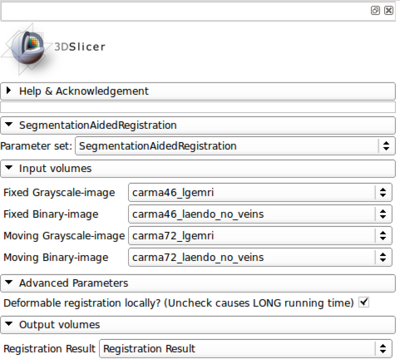
- Step 1. Give the input images, including:
- Fixed grayscale image
- Segmentation label image for the target region in the fixed image
- Moving grayscale image
- Segmentation label image for the target region in the moving image
- Step 2. Give the output filename.
- Run
- There is an optional parameter:
- Only perform deformable registration locally? Yes by default. The output registered grayscale image will only be around the target region. Uncheck to get the registered moving image in the entire region, but this is VERY SLOW.
Panels and their use
- Module panel
- Input
- Fixed grayscale image
- Segmentation label image for the target region in the fixed image
- Moving grayscale image
- Segmentation label image for the target region in the moving image
- Output
- Registered moving image
- Parameter (pptional)
- Only perform deformable registration locally? Yes by default. The output registered grayscale image will only be around the target region. Uncheck to get the registered moving image in the entire region, but this is VERY SLOW.
Similar Modules
References
- Y Gao, Y Rathi, S Bouix, A Tannenbaum; Filtering in the Diffeomorphism Group and the Registration of Point Sets; IEEE Transactions on Image Processing 21 (10), 4383--4396
- Y Gao, B Gholami, RS MacLeod, J Blauer, WM Haddad, A Tannenbaum; Segmentation of the Endocardial Wall of the Left Atrium using Local Region-Based Active Contours and Statistical Shape Learning. Proceedings of SPIE Medical Imaging 2010.
Information for Developers
| Section under construction. |