Documentation/Nightly/Modules/MultiVolumeImporter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is supported by NA-MIC, NAC, NCIGT, and the Slicer Community. This work is partially supported by the following grants: P41EB015898, P41RR019703, R01CA111288 and U01CA151261. | |||||||||||
|
Module Description
This module provides support for importing multivolume (multiframe) data.
Use Cases
Most frequently used for these scenarios:
- importing multiple frames defined in the same coordinate frame, saved as individual volumes in NRRD, NIfTI, or any other image format supported by 3D Slicer
- for the multi-frame data stored in DICOM format, DICOM module should be used instead; the data should be automatically recognized while parsing the dataset in DICOM module.
Tutorials
- Sample datasets are available:
- File:Cardiac ECGg CT.tgz (ECG-gated contrast-enhanced cardiac CT). Short movie
Panels and their use
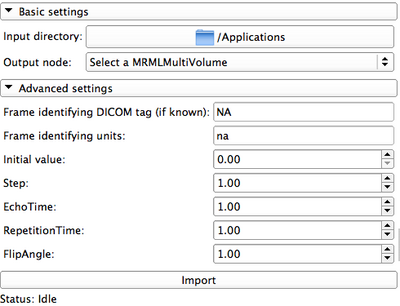
|
- Advanced settings: contains elements that can be changed by the user. These items will be associated with the resulting multivolume, and will be available in case they are needed for the subsequent post-processing of the data (e.g., for pharmacokinetic modeling)
- DICOM tag: in all modes, shows the DICOM tag that will be used to separate individual frames/volumes in the DICOM series. This field does not have meaning when the input data type is non-DICOM!
- Frame identifying units: automatically populated for pre-defined tags. Needs to be defined for other input data types.
- Initial value: values of the frame-identifying units for non-DICOM data type.
- Import button: once the panels are populated with the appropriate settings, hit this button to import the dataset into Slicer. Note that depending on the size of the data this operation can take significant time, so be patient!
Related Modules
Once the multivolume dataset is loaded, it can be viewed and further explored using MultiVolumeExplorer module.
Documentation/Nightly/Modules/PkModeling extension can be used for pharmacokinetic analysis of the DCE MRI data.
References
- Development of this module was initiated at the 2012 NA-MIC Project week at SLC (see http://wiki.na-mic.org/Wiki/index.php/2012_Project_Week:4DImageSlicer4)
Information for Developers
This module is an external Slicer module. The source code is available on Github here: https://github.com/fedorov/MultiVolumeImporter