Difference between revisions of "Documentation/Nightly/Modules/MeshCompression"
(changed logo) |
|||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | Extension: [[Documentation/{{documentation/version}}/Extensions/ | + | Extension: [[Documentation/{{documentation/version}}/Extensions/CBC_3D_I2MConversion |CBC 3D I2MConversion]]<br> |
Acknowledgments: | Acknowledgments: | ||
| − | *This work was partially supported by NIH R44 OD018334-03A, grant CCF- | + | *This work was partially supported by NIH R44 OD018334-03A, grant CCF-1439079 and by the Richard T.Cheng Endowment. |
Author: Fotis Drakopoulos<br> | Author: Fotis Drakopoulos<br> | ||
| − | Contributors: Fotis Drakopoulos (CRTC), Yixun Liu (CRTC), Andrey Fedorov (CRTC), Nikos Chrisochoides (CRTC)<br> | + | Contributors: Joi Best (CRTC), Fotis Drakopoulos (CRTC), Yixun Liu (CRTC), Andrey Fedorov (CRTC), Nikos Chrisochoides (CRTC)<br> |
| − | Contact: Nikos Chrisochoides | + | Contact: Nikos Chrisochoides: <email>npchris@gmail.com</email> , Joi Best: <email>jbest002@odu.edu</email><br> |
| − | Website: https://crtc.cs.odu.edu | + | Website: https://crtc.cs.odu.edu <br> |
License: BSD<br> | License: BSD<br> | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |Image: | + | |Image:Crtc logo.png|Center for Real-time Computing |
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 49: | Line 49: | ||
{| | {| | ||
| − | |[[File:target_points.jpg|thumb| | + | |[[File:target_points.jpg|thumb|960px|How the non-connectivity parameter influences the number of the extracted target points]] |
|} | |} | ||
| Line 75: | Line 75: | ||
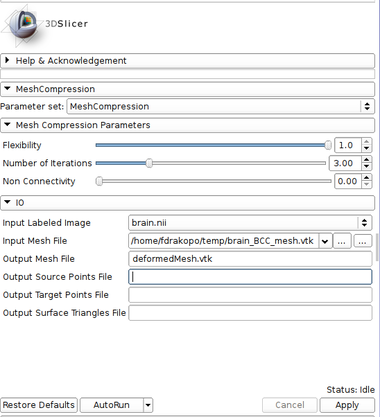
|[[Image:MCPanel.jpg|thumb|380px|User Interface]] | |[[Image:MCPanel.jpg|thumb|380px|User Interface]] | ||
|} | |} | ||
| + | |||
| + | '''Note: this module will only work if the input image has identity orientation!''' | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 14:08, 7 August 2018
Home < Documentation < Nightly < Modules < MeshCompression
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: CBC 3D I2MConversion
Author: Fotis Drakopoulos | |||
|
Module Description
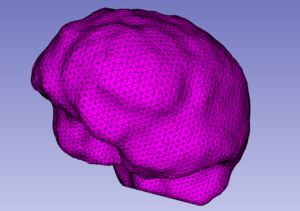
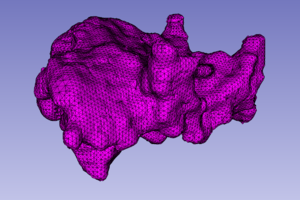
The Mesh Compression (MC) module deforms the input tetrahedral mesh towards the boundaries of the input labeled image. Two point sets are extracted for the mesh deformation. The first (source point set) consists of the surface vertices in the input mesh. The second (target point set) consists of the surface edge points in the input labeled image. After the extraction of the two point sets, the input mesh is deformed by registering the source to the target point set using a Physics-Based Non-Rigid Registration method. The current version of the module supports a single-tissue labeled image and mesh.
Use Cases
Tutorials
N/A
Panels and their use
|
Note: this module will only work if the input image has identity orientation!
Similar Modules
References
- Yixun Liu, Panagiotis Foteinos, Andrey Chernikov and Nikos Chrisochoides, "Mesh Deformation-based Multi-tissue Mesh Generation for Brain Images", Engineering with Computers, Volume 28, pages 305-318, 2012.
Information for Developers
| Section under construction. |