Difference between revisions of "Documentation/Nightly/Modules/LSContrastEnhancer"
From Slicer Wiki
Acsenrafilho (talk | contribs) m |
Acsenrafilho (talk | contribs) m |
||
| Line 22: | Line 22: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This | + | [[Image:LSContrastEnhancer-icon.png|right]] |
| + | |||
| + | This Module offer a method to increase hyperintense lesions on T2-FLAIR MRI acquisitions, which is mainly applicable in Multiple Sclerosis lesion detection. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | * Use Case 1: | + | * Use Case 1: Increase contrast in abnormal voxels in white matter |
| − | ** | + | **There are some lesion segmentation approaches that relies on the voxel intensity level presented in the lesion signal, where the LS Contrast Enhancer module can be helpful to increase the contrast between lesions and surrounding brain tissues (mainly White Matter) |
| − | <gallery widths=" | + | |
| − | Image: | + | <gallery widths="300px" heights="300px" perrow="3"> |
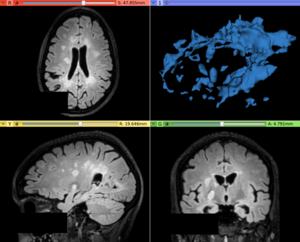
| − | Image: | + | Image:T2FLAIR_beforeContrast.png|Input T2-FLAIR image with Multiple Sclerosis lesions |
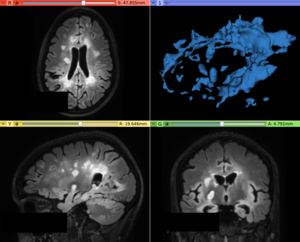
| + | Image:T2FLAIR_afterContrast.png|Output image with the hyperintense lesions enhanced in contrast with the surrounding tissues | ||
</gallery> | </gallery> | ||
| Line 38: | Line 41: | ||
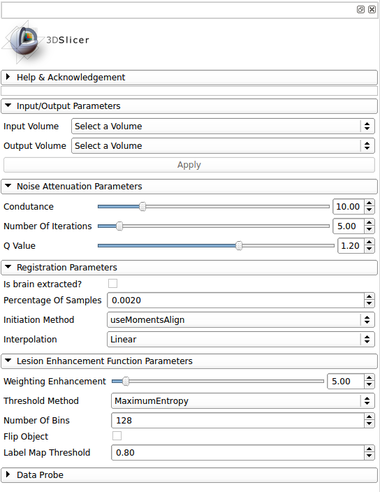
[[Image:lscontrastenhancer_gui.png|thumb|380px|User Interface]] | [[Image:lscontrastenhancer_gui.png|thumb|380px|User Interface]] | ||
| − | + | Input/Output Parameters: | |
*Input Volume | *Input Volume | ||
| − | |||
*Output Volume | *Output Volume | ||
| − | ** | + | **Output enhanced volume |
| − | + | Noise Attenuation Parameters: | |
*Conductance | *Conductance | ||
**The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space. | **The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space. | ||
| − | * | + | *Number Of Iterations |
| + | **The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter | ||
| + | *Q Value | ||
| + | **The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. See the reference paper[1] to choose the appropriate q value (at moment, only tested in MRI T1 and T2 weighted images) | ||
| + | |||
| + | Registration Parameters: (based on [https://www.slicer.org/wiki/Documentation/Nightly/Modules/BRAINSFit BRAINSFit] module) | ||
| + | *Is brain extracted | ||
| + | **Is the input data (T1 and T2-FLAIR) already brain extracted? | ||
| + | *Percentage Of Samples | ||
| + | **Percentage of voxel used in registration | ||
| + | *Initiation Method | ||
| + | **Initialization method used for the MNI152 registration | ||
| + | *Interpolation | ||
| + | **Choose the interpolation method used to register the standard space to input image space. Options: Linear, NearestNeighbor, B-Spline | ||
| + | |||
| + | Lesion Enhancement Function Parameters: | ||
| + | *Weighting Enhancement | ||
| + | **Weighting enhancement value, in percentage, that will be used to increase the lesion signal in the image | ||
| + | *Threshold Method | ||
| + | **Choose the threhsold method for the lesion enhancement procedure. Options: MaximumEntropy, Otsu, Moments, Intermodes and IsoData | ||
| + | *Number Of Bins | ||
| + | **Number Of Bins for the histogram calculation | ||
| + | *Flip Object | ||
| + | **Flip object in the image. This inform if the dark part of the histogram that should be enhanced | ||
| + | *Label Map Threshold | ||
| + | **Threshold for the lesion label map | ||
| + | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 15:53, 9 December 2016
Home < Documentation < Nightly < Modules < LSContrastEnhancer
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: LesionSpotlight | |||||||
|
Module Description
This Module offer a method to increase hyperintense lesions on T2-FLAIR MRI acquisitions, which is mainly applicable in Multiple Sclerosis lesion detection.
Use Cases
- Use Case 1: Increase contrast in abnormal voxels in white matter
- There are some lesion segmentation approaches that relies on the voxel intensity level presented in the lesion signal, where the LS Contrast Enhancer module can be helpful to increase the contrast between lesions and surrounding brain tissues (mainly White Matter)
Panels and their use
Input/Output Parameters:
- Input Volume
- Output Volume
- Output enhanced volume
Noise Attenuation Parameters:
- Conductance
- The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space.
- Number Of Iterations
- The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter
- Q Value
- The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. See the reference paper[1] to choose the appropriate q value (at moment, only tested in MRI T1 and T2 weighted images)
Registration Parameters: (based on BRAINSFit module)
- Is brain extracted
- Is the input data (T1 and T2-FLAIR) already brain extracted?
- Percentage Of Samples
- Percentage of voxel used in registration
- Initiation Method
- Initialization method used for the MNI152 registration
- Interpolation
- Choose the interpolation method used to register the standard space to input image space. Options: Linear, NearestNeighbor, B-Spline
Lesion Enhancement Function Parameters:
- Weighting Enhancement
- Weighting enhancement value, in percentage, that will be used to increase the lesion signal in the image
- Threshold Method
- Choose the threhsold method for the lesion enhancement procedure. Options: MaximumEntropy, Otsu, Moments, Intermodes and IsoData
- Number Of Bins
- Number Of Bins for the histogram calculation
- Flip Object
- Flip object in the image. This inform if the dark part of the histogram that should be enhanced
- Label Map Threshold
- Threshold for the lesion label map
Similar Modules
References
- paper
Information for Developers
| Section under construction. |