Difference between revisions of "Documentation/Nightly/Modules/FastGrowCut"
Liangjiazhu (talk | contribs) |
Liangjiazhu (talk | contribs) |
||
| Line 135: | Line 135: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | * Liangjia Zhu, Ivan Kolesov, Yi Gao, Ron Kikinis, Allen Tannenbaum. An Effective Interactive Medical Image Segmentation Method Using Fast GrowCut, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Interactive Medical Image Computing Workshop, 2014 | + | * Liangjia Zhu, Ivan Kolesov, Yi Gao, Ron Kikinis, Allen Tannenbaum. An Effective Interactive Medical Image Segmentation Method Using Fast GrowCut, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Interactive Medical Image Computing Workshop, 2014. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
Revision as of 20:18, 5 April 2015
Home < Documentation < Nightly < Modules < FastGrowCut
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant U54 EB005149. Information on NA-MIC can be obtained from the NA-MIC website. Author: Liangjia Zhu, Stony Brook University
|
|
|
Module Description
This is a fast implementation of the GrowCut method. It supports multi-label segmentation and user online interactions. Please see the references below for more details.
Tutorials
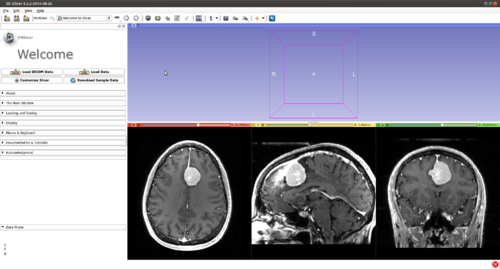
Step 1.) Add data volume to segment
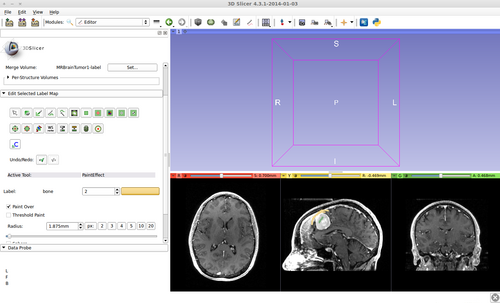
Step 2.) Go to the “Editor” module, select the volume loaded in Step 1 as the “Master Volume” in the “Create and Select Label Maps” drop-down menu
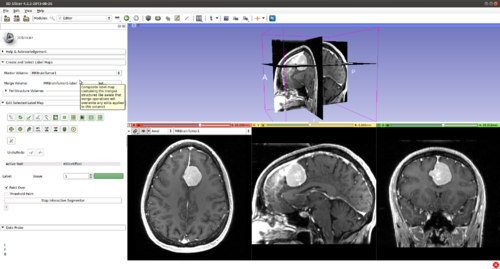
Step 3.) Select the “FastGrowCutEffect” effect in the “Edit Selected Label Map” drop-down menu
Step 4.) Press the “Start Fast GrowCut” button ( FastGrowCut is now running in the background until the “Stop FastGrowCut” button is pressed)
Step 5.) Turn “On” all three slice views in the 3D Plane
Step 6.) Initialize the segmentation using fast GrowCut
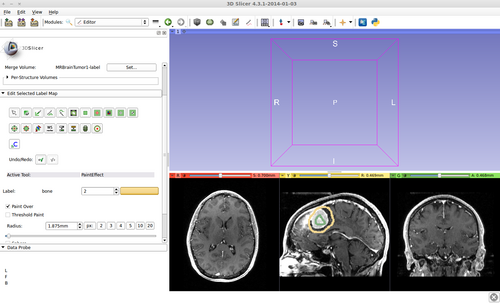
- (a) go to PaintEffect to draw seed regions (label 1 for foreground and 2 for background), then press 'G' to run fast GrowCut.
- (b) If not satified, press 'S' to toggle between seed image and segmentation result. Edit on the seed image to reduce over/under segmentaions.
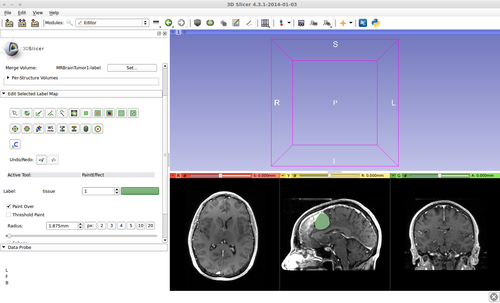
- (c) Once finished editing on the seed image, press 'G' to run fast GrowCut again.
The steps 6 (b) and (c) may be repeated a couple of times until satisfied.
Multi-Label Segmentation Examples
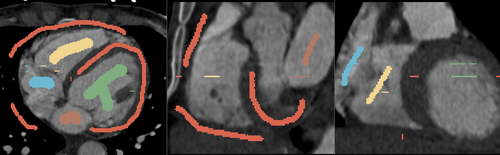
The module supports multi-label segmentations. Two examples are shown below.
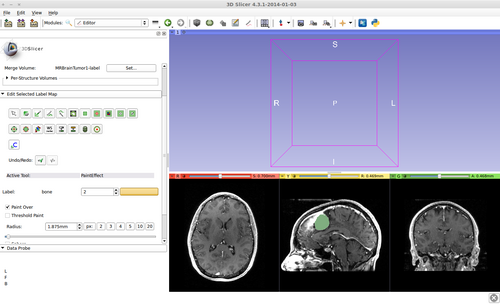
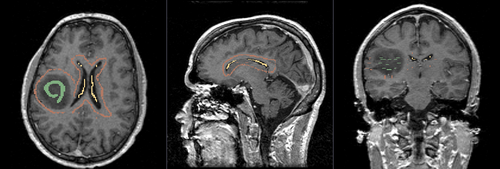
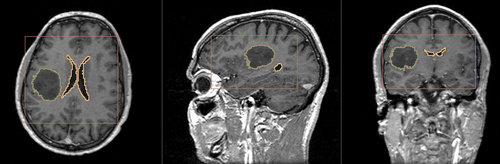
- Brain ventricle and tumor segmentation
1) Seed image
2) Segmentation results
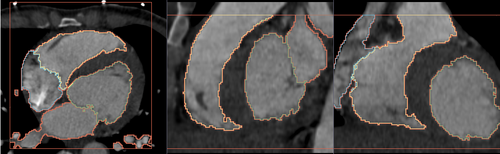
- Heart chamber segmentation
1) Seed image
2) Segmentation results
Similar Modules
References
- Liangjia Zhu, Ivan Kolesov, Yi Gao, Ron Kikinis, Allen Tannenbaum. An Effective Interactive Medical Image Segmentation Method Using Fast GrowCut, International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Interactive Medical Image Computing Workshop, 2014.
Information for Developers
| Section under construction. |