Difference between revisions of "Documentation/Nightly/Modules/DeepInfer"
| (4 intermediate revisions by the same user not shown) | |||
| Line 26: | Line 26: | ||
[[File:Deepinfer-256.png|128px|]] | [[File:Deepinfer-256.png|128px|]] | ||
<br> | <br> | ||
| − | DeepInfer is an | + | DeepInfer is an open-source deep learning deployment toolkit for medical imaging and image-guided therapy. With DeepInfer 3D Slicer users can deploy trained |
deep learning and machine learning models in their workflow. Also, DeepInfer allows machine learning experts to package, | deep learning and machine learning models in their workflow. Also, DeepInfer allows machine learning experts to package, | ||
ship, and deploy their models, thereby additionally allowing clinical researchers and biomedical engineers to use | ship, and deploy their models, thereby additionally allowing clinical researchers and biomedical engineers to use | ||
| Line 36: | Line 36: | ||
Docker for Windows requires Windows 10 Pro (64bit) and Microsoft Hyper-V. You also need to enable virtualization in BIOS settings. | Docker for Windows requires Windows 10 Pro (64bit) and Microsoft Hyper-V. You also need to enable virtualization in BIOS settings. | ||
* [https://docs.docker.com/docker-for-windows/install/ Docker installation guide for Windows] | * [https://docs.docker.com/docker-for-windows/install/ Docker installation guide for Windows] | ||
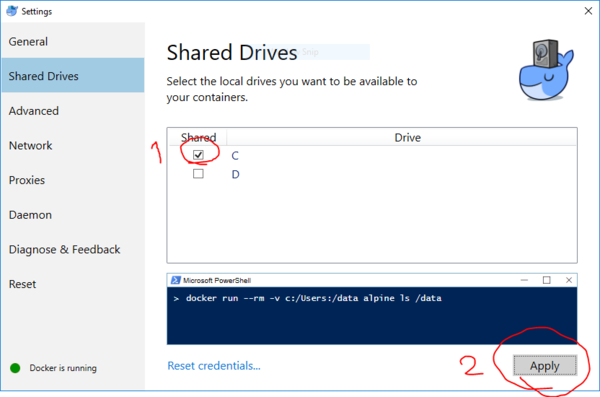
| − | * Share drive C with docker containers from settings. | + | * VERY IMPORTANT: Share drive C with docker containers from settings. |
| + | [[File:DockerWindows.PNG|600px|]]<br><br> | ||
==== Mac OS X ==== | ==== Mac OS X ==== | ||
| Line 45: | Line 46: | ||
* Give non-root access to docker using this [http://askubuntu.com/a/477554 guide]. | * Give non-root access to docker using this [http://askubuntu.com/a/477554 guide]. | ||
| − | {{documentation/{{documentation/version}}/module-section| | + | {{documentation/{{documentation/version}}/module-section|Demo}} |
| + | * [https://www.youtube.com/watch?v=7k_SLtmM2Kw Demo Video] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
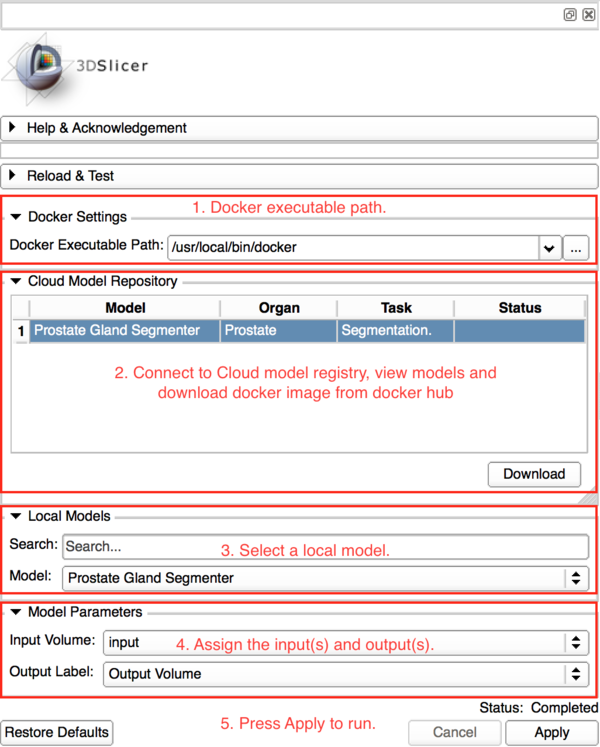
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | + | [[File:Deepinfer-panels.png|600px|]] | |
{{documentation/{{documentation/version}}/module-section|Deployed Models}} | {{documentation/{{documentation/version}}/module-section|Deployed Models}} | ||
Latest revision as of 00:36, 12 August 2017
Home < Documentation < Nightly < Modules < DeepInfer
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: DeepInfer
|
Module Description

DeepInfer is an open-source deep learning deployment toolkit for medical imaging and image-guided therapy. With DeepInfer 3D Slicer users can deploy trained
deep learning and machine learning models in their workflow. Also, DeepInfer allows machine learning experts to package,
ship, and deploy their models, thereby additionally allowing clinical researchers and biomedical engineers to use
task-specific deep models, without the need for further software development and configuration.
Setup Guide
In order to use DeepInfer, Docker is required to be installed and configured properly.
Windows
Docker for Windows requires Windows 10 Pro (64bit) and Microsoft Hyper-V. You also need to enable virtualization in BIOS settings.
- Docker installation guide for Windows
- VERY IMPORTANT: Share drive C with docker containers from settings.
Mac OS X
Ubuntu
- Docker installation guide for Ubuntu
- Give non-root access to docker using this guide.
Demo
Panels and their use
Deployed Models
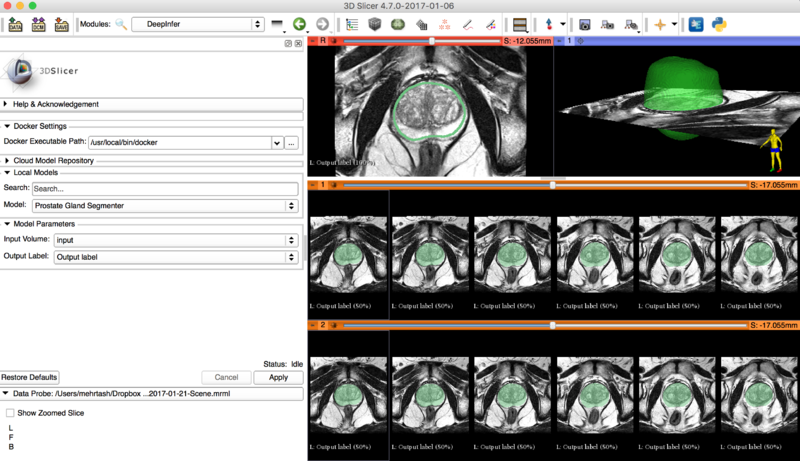
Prostate Segmentation in Targeted MRI-Guided Biopsy
This model is trained for the task of rough delineation of the prostate gland that is a necessary step in MRI-guided targeted biopsy. The deployed model is trained on transaxial T2-weighted MRIs of the prostate (without endorectal coil on a 3T MRI machine) collected during MR-guided prostate biopsy procedures. The network based on a customized variant of the U-Net architecture. The network is trained on N = 224 patients on a total number of 26250 2D slices of prostate images and achieved 76.25% accuracy on N = 57 validation patients (2184 2D slices).
Citing DeepInfer
To acknowledge DeepInfer and 3D Slicer, please cite the please cite the DeepInfer web site and following publications when publishing work that uses or incorporates DeepInfer and 3D Slicer:
- Mehrtash, A., Pesteie, M., Hetherington, J., Behringer, P.A., Kapur, T., Wells, W.M., Rohling, R., Fedorov, A. and Abolmaesumi, P., 2017, March. DeepInfer: open-source deep learning deployment toolkit for image-guided therapy. In SPIE Medical Imaging (pp. 101351K-101351K). International Society for Optics and Photonics.
- Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J-C., Pujol S., Bauer C., Jennings D., Fennessy F.M., Sonka M., Buatti J., Aylward S.R., Miller J.V., Pieper S., Kikinis R. 3D Slicer as an Image Computing Platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012 Nov;30(9):1323-41. PMID: 22770690. PMCID: PMC3466397.
Information for Developers
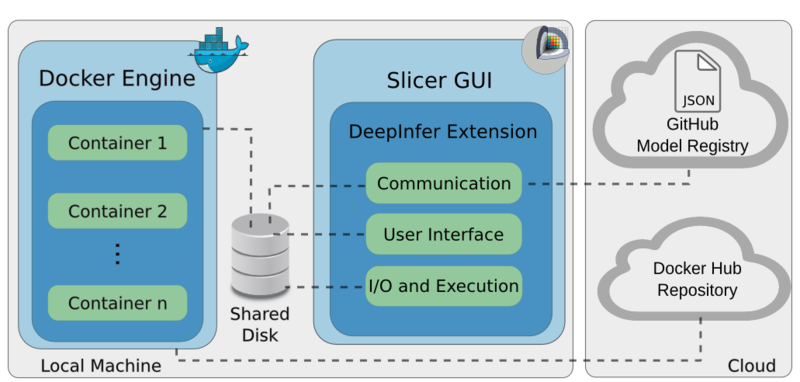
Architecture
The figure below shows the architecture of the DeepInfer toolkit, which has three components: the Docker engine,
the DeepInfer 3D Slicer extension [1], and the cloud model registry [2].
The Docker engine consists of local Docker containers that include the deployed models, as well as all of the required deep learning frameworks, which
process incoming data and produce respective results.
Data streaming between the Docker engine and the GUI is achieved by sharing a local folder with the specific Docker image.
Packaging and Deployment Example
Prostate Segmenter Repository is an example of the code that can be used as a reference to package and deploy a model in DeepInfer. It shows how you can build a docker and package trained weights inside.