Difference between revisions of "Documentation/Nightly/Modules/Calculator"
From Slicer Wiki
(Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | + | ||
| − | + | <br> | |
| − | + | '''Authors:''' Frédéric Panthier, Lounès Illoul, Laurent Berthe, Steeve Doizi, Oliver Traxer | |
| − | + | ||
| − | Contact: | + | Contact: Frédéric Panthier, <email>fredericpanthier@gmail.com</email><br> |
| + | |||
| + | '''License:''' [https://github.com/Slicer/Slicer/blob/master/License.txt Slicer License] | ||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | |||
| − | |||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | |||
<!-- | <!-- | ||
Here comes a description what the module is good for. Explain briefly how it works and point to the [[documentation/{{documentation/version}}/Modules/{{documentation/modulename}}#References|references]] giving more details on the algorithm. | Here comes a description what the module is good for. Explain briefly how it works and point to the [[documentation/{{documentation/version}}/Modules/{{documentation/modulename}}#References|references]] giving more details on the algorithm. | ||
| − | |||
| − | |||
{{documentation/{{documentation/version}}/module-description}} | {{documentation/{{documentation/version}}/module-description}} | ||
--> | --> | ||
| + | {| | ||
| + | |[[File:calculator.png|thumb|left]] | ||
| + | |“Calculator” consists in a volumetric evaluation of kidney stones and also provides information on surgical duration. Moreover, it provides the estimated time of lithotripsy and consequently the operative time. It provides in a few seconds a 3D view of the stone(s) and axial, coronal and sagittal views with CT-scan DICOMs. | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 37: | Line 37: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| − | + | ||
| + | Below, we describe the Instructions Guide to use Calculator: | ||
| + | |||
| + | # Open 3DSlicer and download “Calculator” using the “Extension Manager” | ||
| + | # Import DICOM data from a CT-scan (better with non-enhanced series) | ||
| + | # Open “Crop Volume” module | ||
| + | #* “Create a new annotation ROI” (Region Of Interest) | ||
| + | #* Size the ROI in x, y and z axes using axial (red), sagittal (yellow) and coronal (green) view in four-up disposition. | ||
| + | #* Create a new volume (name will be: “name cropped”) | ||
| + | #* Click “Apply” button | ||
| + | # Open “Calculator” module: | ||
| + | #* Select volume: “name cropped” | ||
| + | #* Choose threshold (houndfields units) minimum and maximun to fit to the stone | ||
| + | #* If there are multiples stones select “split islands into segments” with a recommended “minimum size” of 40 voxels | ||
| + | #* Select a mode of treatment (laser source, core-diameter of laser fiber, laser settings and stone type) | ||
| + | #* Select “show 3D” if you want to visualize in 3D-view the segmented stone(s) | ||
| + | #* Click “Apply” button. A table will appear to show the segment’s name, volume (mm3) and time of lithotripsy (min) | ||
| + | |||
| + | {| | ||
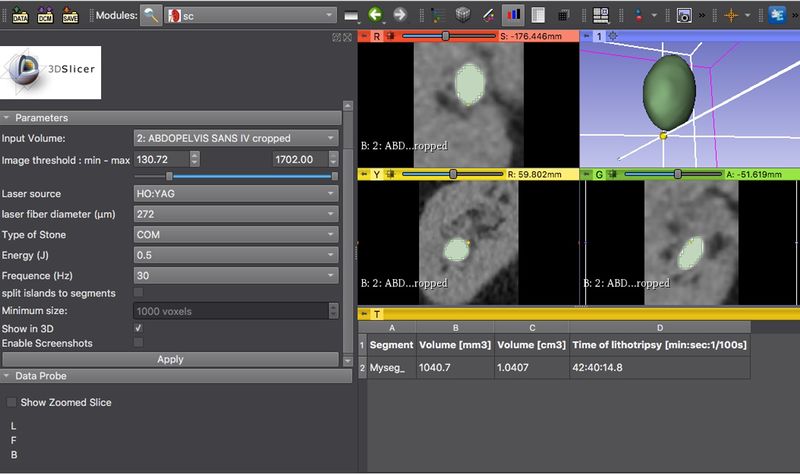
| + | |[[File:Calculator-Screenshot.jpg|800px|thumb|left|Screenshot]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Latest revision as of 16:51, 7 November 2019
Home < Documentation < Nightly < Modules < Calculator
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Contact: Frédéric Panthier, <email>fredericpanthier@gmail.com</email> License: Slicer License |
Module Description
| “Calculator” consists in a volumetric evaluation of kidney stones and also provides information on surgical duration. Moreover, it provides the estimated time of lithotripsy and consequently the operative time. It provides in a few seconds a 3D view of the stone(s) and axial, coronal and sagittal views with CT-scan DICOMs. |
Use Cases
N/A
Tutorials
Below, we describe the Instructions Guide to use Calculator:
- Open 3DSlicer and download “Calculator” using the “Extension Manager”
- Import DICOM data from a CT-scan (better with non-enhanced series)
- Open “Crop Volume” module
- “Create a new annotation ROI” (Region Of Interest)
- Size the ROI in x, y and z axes using axial (red), sagittal (yellow) and coronal (green) view in four-up disposition.
- Create a new volume (name will be: “name cropped”)
- Click “Apply” button
- Open “Calculator” module:
- Select volume: “name cropped”
- Choose threshold (houndfields units) minimum and maximun to fit to the stone
- If there are multiples stones select “split islands into segments” with a recommended “minimum size” of 40 voxels
- Select a mode of treatment (laser source, core-diameter of laser fiber, laser settings and stone type)
- Select “show 3D” if you want to visualize in 3D-view the segmented stone(s)
- Click “Apply” button. A table will appear to show the segment’s name, volume (mm3) and time of lithotripsy (min)
Panels and their use
N/A
Similar Modules
N/A
References
N/A
Information for Developers
| Section under construction. |