Difference between revisions of "Documentation/Nightly/Modules/BrainVolumeRefinement"
From Slicer Wiki
Acsenrafilho (talk | contribs) (Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
Acsenrafilho (talk | contribs) (ENH: Improved the general information of BVeR module) |
||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | Extension: [[Documentation/{{documentation/version}}/Extensions/ | + | Extension: [[Documentation/{{documentation/version}}/Extensions/BabyBrain|Baby Brain Toolkit]]<br> |
Webpage: http://dcm.ffclrp.usp.br/csim/<br> | Webpage: http://dcm.ffclrp.usp.br/csim/<br> | ||
| − | Author: Antonio Carlos da S. Senra Filho, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics)<br> | + | Author: Antonio Carlos da S. Senra Filho and Fabricio Henrique Simozo, CSIM Laboratory (University of Sao Paulo, Department of Computing and Mathematics)<br> |
Contact: Antonio Carlos da S. Senra Filho, <email>acsenrafilho@usp.br</email><br> | Contact: Antonio Carlos da S. Senra Filho, <email>acsenrafilho@usp.br</email><br> | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| Line 16: | Line 16: | ||
|Image:CSIM-logo.png|CSIM Laboratory | |Image:CSIM-logo.png|CSIM Laboratory | ||
|Image:USP-logo.png|University of Sao Paulo | |Image:USP-logo.png|University of Sao Paulo | ||
| − | |Image: | + | |Image:CNPq-logo.png|CNPq Brazil |
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 22: | Line 22: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module | + | This is a CLI module for Brain Volume Refinement (BVeR) tool, which is useful to correct brain segmentation errors that may appear in commonly brain extraction methods. The BVeR algorithm was extensively tested on structural MRI images (T1 and T2) of normal individuals. Further details are found at '''PAPER''' |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | * Use Case 1: | + | * Use Case 1: Segmentation outliers removal from previous brain extraction process |
| − | ** | + | **It is frequent to appear segmentation errors (outliers) in commonly used brain extraction algorithms (BET, FreeSufer, BSE, AFNI, ROBEX, etc), thus the BVeR application can be helpful. |
| − | * Use Case 2: | + | * Use Case 2: Improving brain morphological measurement in large-scale studies |
| − | ** | + | **Due to decrease of segmentation errors, the outcome of large-scale morphological measurements can have better precision (e.g. cortical thickness and brain atrophy) |
| − | |||
| − | |||
<gallery widths="300px" perrow="3"> | <gallery widths="300px" perrow="3"> | ||
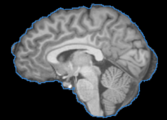
| − | Image: | + | Image:T1_BET.png|A T1 weighted MRI image with the brain extracted using BET. |
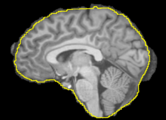
| − | Image: | + | Image:T1_BET_BVeR.png|The BVeR application showing segmentation outlier corrections (brain external frontier) |
</gallery> | </gallery> | ||
| Line 42: | Line 40: | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
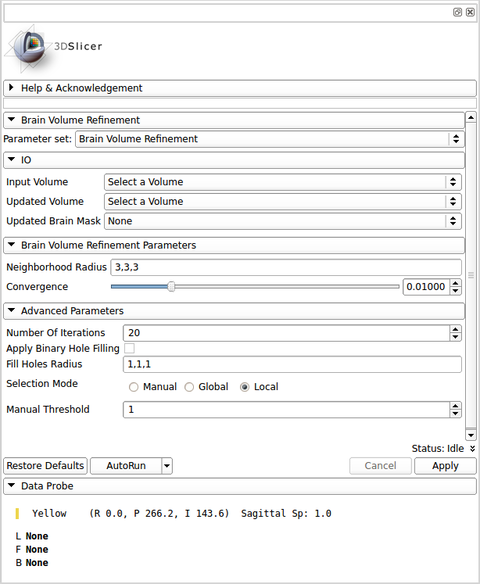
| − | [[Image: | + | [[Image:BVeR_gui.png|thumb|480px|User Interface]] |
'''IO:''' | '''IO:''' | ||
*'''Input Volume''' | *'''Input Volume''' | ||
| − | ** | + | **This is a previous brain extracted image which presents small segmentation outliers on the brain frontier (e.g. dura matter or bone marrow). |
| − | *''' | + | *'''Updated Volume''' |
| − | ** | + | **Updated Volume with the outlier segmentation being corrected. |
| + | *'''Updated Brain Mask''' | ||
| + | **Updated brain mask obtained from the output image. | ||
| − | ''' | + | '''Brain Volume Refinement Parameters:''' |
| − | *''' | + | *'''Neighbourhood Radius''' |
| − | ** | + | **A list of 3 values indicating the (x,y,z) size of the neighbourhood. This should large enough in order to get a consistent local statistics (values around 3 to 8). Example: a radius of (1,1,1) creates a neighbourhood of (3,3,3) in image space. |
| − | *''' | + | *'''Convergence''' |
| − | ** | + | **A relative value that indicates how permissive is the algorithm to keep changing the brain borders. This counts how many voxels were changed in previous iterations and then estimate whether the total amount of changes reaches a limit (regarding the total number of voxels changed in the brain volume). High values will result in conservative outputs (fewer changes in the brain volume), on the other hand, low values will force a strong volume difference. |
| − | *''' | + | |
| − | ** | + | '''Advanced Parameters''' |
| − | *''' | + | *'''Number Of Iterations''' |
| − | ** | + | **Maximum number of iterations. The brain mask is iteratively updated in order to vanish bigger error in the previous brain extraction result. This is an up limit threshold in order to avoid infinite loop in the brain volume correction. If the convergence level does not reach a stable result, then the number of iteration limit will stop the algorithm. |
| − | *''' | + | *'''Apply Binary Hole Filling''' |
| − | ** | + | **Choose if you want to fill holes in the first brain mask. This is important to not place a searching window inside the brain area. One can avoid this if a previous visual check was made in order to confirm that there are no zero values inside the brain area. |
| − | *''' | + | *'''Fill Holes Radius''' |
| − | **The | + | **A list of 3 values indicating the (x,y,z) size of the neighbourhood used in the binary filling hole procedure. This parameter is only used if "Apply Binary Hole Filling" option is checked. Example: a radius of 1 creates an isotropic neighbourhood of (3,3,3). |
| + | *'''Selection Mode''' | ||
| + | **The BVeR algorithm relies on the estimate of non-brain voxels that are still present in the input image. These outliers voxels are detected by a determined grey level threshold, which is iteratively updated. This option will define what is the type of the grey level threshold used in the entire process. | ||
| + | ***Local = The threshold is calculated for each fixed neighbourhood | ||
| + | ***Global=The threshold is set by the global image mean value (zeros are not considered) | ||
| + | ***Manual=The user defines a fixed threshold. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | + | N/A | |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | * | + | * PAPER |
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 14:15, 7 March 2018
Home < Documentation < Nightly < Modules < BrainVolumeRefinement
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: Baby Brain Toolkit | |||||||
|
Module Description
This is a CLI module for Brain Volume Refinement (BVeR) tool, which is useful to correct brain segmentation errors that may appear in commonly brain extraction methods. The BVeR algorithm was extensively tested on structural MRI images (T1 and T2) of normal individuals. Further details are found at PAPER
Use Cases
- Use Case 1: Segmentation outliers removal from previous brain extraction process
- It is frequent to appear segmentation errors (outliers) in commonly used brain extraction algorithms (BET, FreeSufer, BSE, AFNI, ROBEX, etc), thus the BVeR application can be helpful.
- Use Case 2: Improving brain morphological measurement in large-scale studies
- Due to decrease of segmentation errors, the outcome of large-scale morphological measurements can have better precision (e.g. cortical thickness and brain atrophy)
Panels and their use
IO:
- Input Volume
- This is a previous brain extracted image which presents small segmentation outliers on the brain frontier (e.g. dura matter or bone marrow).
- Updated Volume
- Updated Volume with the outlier segmentation being corrected.
- Updated Brain Mask
- Updated brain mask obtained from the output image.
Brain Volume Refinement Parameters:
- Neighbourhood Radius
- A list of 3 values indicating the (x,y,z) size of the neighbourhood. This should large enough in order to get a consistent local statistics (values around 3 to 8). Example: a radius of (1,1,1) creates a neighbourhood of (3,3,3) in image space.
- Convergence
- A relative value that indicates how permissive is the algorithm to keep changing the brain borders. This counts how many voxels were changed in previous iterations and then estimate whether the total amount of changes reaches a limit (regarding the total number of voxels changed in the brain volume). High values will result in conservative outputs (fewer changes in the brain volume), on the other hand, low values will force a strong volume difference.
Advanced Parameters
- Number Of Iterations
- Maximum number of iterations. The brain mask is iteratively updated in order to vanish bigger error in the previous brain extraction result. This is an up limit threshold in order to avoid infinite loop in the brain volume correction. If the convergence level does not reach a stable result, then the number of iteration limit will stop the algorithm.
- Apply Binary Hole Filling
- Choose if you want to fill holes in the first brain mask. This is important to not place a searching window inside the brain area. One can avoid this if a previous visual check was made in order to confirm that there are no zero values inside the brain area.
- Fill Holes Radius
- A list of 3 values indicating the (x,y,z) size of the neighbourhood used in the binary filling hole procedure. This parameter is only used if "Apply Binary Hole Filling" option is checked. Example: a radius of 1 creates an isotropic neighbourhood of (3,3,3).
- Selection Mode
- The BVeR algorithm relies on the estimate of non-brain voxels that are still present in the input image. These outliers voxels are detected by a determined grey level threshold, which is iteratively updated. This option will define what is the type of the grey level threshold used in the entire process.
- Local = The threshold is calculated for each fixed neighbourhood
- Global=The threshold is set by the global image mean value (zeros are not considered)
- Manual=The user defines a fixed threshold.
- The BVeR algorithm relies on the estimate of non-brain voxels that are still present in the input image. These outliers voxels are detected by a determined grey level threshold, which is iteratively updated. This option will define what is the type of the grey level threshold used in the entire process.
Similar Modules
N/A
References
- PAPER
Information for Developers
| Section under construction. |