Difference between revisions of "Documentation/Nightly/Modules/BrainStructuresSegmenter"
Acsenrafilho (talk | contribs) (BasicTissueSegmenter wikipage created) |
Acsenrafilho (talk | contribs) m |
||
| Line 22: | Line 22: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module offer a . | + | This module offer a general brain segmentation pipeline, where it call different algorithm depending the tissue type that the user wants to segment. At this moment, the basic white matter, gray matter and CSF tissues segmentation algorithm is presented, however, more complex segmentation algorithms will be added in order to evaluate deep gray matter segmentation and others brain tissues types. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | * Use Case 1: a | + | * Use Case 1: Tissue classification |
| − | ** | + | **There are several image quantitative approaches that are applied only in a certain tissue type (for instance, cortical thickness) in which a previous brain segmentation could be needed. |
| + | **A simple brain tissue mask could be obtained from this module (WM, GM and CSF are available at moment). | ||
<gallery widths="200px" perrow="3"> | <gallery widths="200px" perrow="3"> | ||
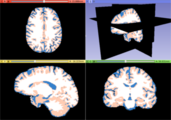
| − | Image: | + | Image:T1_tissues.png|White matter, gray matter and CSF tissues segmented from the previous MRI image |
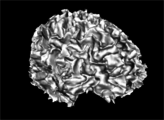
| − | Image: | + | Image:WM_3DReconstruction.png|A white matter mask 3D reconstruction |
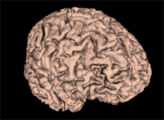
| + | Image:GM_3DReconstruction.png|A gray matter mask 3D reconstruction | ||
</gallery> | </gallery> | ||
| Line 37: | Line 39: | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
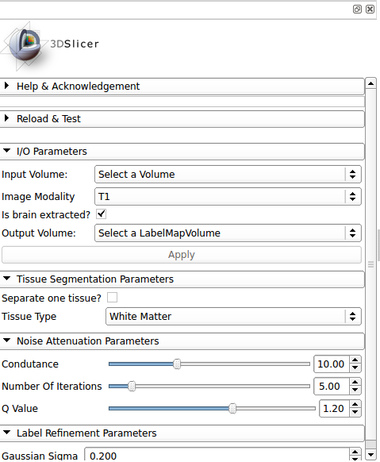
| − | [[Image: | + | [[Image:BrainStructuresSegmenter_gui.png|thumb|380px|User Interface]] |
IO: | IO: | ||
*Input Volume | *Input Volume | ||
| − | ** | + | **Pick the input to the algorithm. This should be an MRI strutural images with a type listed in the Image Modality option |
| + | *Image Modality | ||
| + | **MRI strutural image inserted as a input volume | ||
| + | *Is brain extracted? | ||
| + | **Is the input data already brain extracted? If not, the [https://www.slicer.org/wiki/Documentation/Nightly/Modules/ROBEXBrainExtraction ROBEX] brain extraction method is used | ||
*Output Volume | *Output Volume | ||
| − | ** | + | **Pick the output to the algorithm (a label image) |
| − | + | Tissue Segmentation Parameters: | |
| − | * | + | *Separate one tissue? |
| − | **The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space | + | **Select one tissue type desired to be passed as the output. If checked, the tissue type in Tissue Type option is used |
| − | *Number | + | *Tissue Type |
| − | **The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter | + | **Tissue type that will be resulted from the brain segmentation |
| − | + | ||
| − | + | Noise Attenuation Parameters: | |
| − | * | + | *Condutance |
| − | **The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation. | + | **The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space |
| + | *Number Of Iterations | ||
| + | **The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter | ||
| + | *Q Value | ||
| + | **The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation | ||
| + | |||
| + | Label Refinement Parameters | ||
| + | *Gaussian Sigma | ||
| + | **Label smoothing by a gaussian distribution with variance sigma. The units here is given in mm | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:EMSegmenter-3.6 EM Segmenter] |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | + | N/A | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 18:18, 26 November 2016
Home < Documentation < Nightly < Modules < BrainStructuresSegmenter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: BrainTissuesExtension | |||||||
|
Module Description
This module offer a general brain segmentation pipeline, where it call different algorithm depending the tissue type that the user wants to segment. At this moment, the basic white matter, gray matter and CSF tissues segmentation algorithm is presented, however, more complex segmentation algorithms will be added in order to evaluate deep gray matter segmentation and others brain tissues types.
Use Cases
- Use Case 1: Tissue classification
- There are several image quantitative approaches that are applied only in a certain tissue type (for instance, cortical thickness) in which a previous brain segmentation could be needed.
- A simple brain tissue mask could be obtained from this module (WM, GM and CSF are available at moment).
Panels and their use
IO:
- Input Volume
- Pick the input to the algorithm. This should be an MRI strutural images with a type listed in the Image Modality option
- Image Modality
- MRI strutural image inserted as a input volume
- Is brain extracted?
- Is the input data already brain extracted? If not, the ROBEX brain extraction method is used
- Output Volume
- Pick the output to the algorithm (a label image)
Tissue Segmentation Parameters:
- Separate one tissue?
- Select one tissue type desired to be passed as the output. If checked, the tissue type in Tissue Type option is used
- Tissue Type
- Tissue type that will be resulted from the brain segmentation
Noise Attenuation Parameters:
- Condutance
- The conductance regulates the diffusion intensity in the neighbourhood area. Choose a higher conductance if the input image has strong noise seem in the whole image space
- Number Of Iterations
- The number of iterations regulates the numerical simulation of the anomalous process over the image. This parameters is also related with the de-noising intensity, however it is more sensible to the noise intensity. Choose the higher number of iterations if the image presents high intensity noise which is not well treated by the conductance parameter
- Q Value
- The anomalous parameter (or q value) is the generalization parameters responsible to give the anomalous process approach on the diffusion equation
Label Refinement Parameters
- Gaussian Sigma
- Label smoothing by a gaussian distribution with variance sigma. The units here is given in mm
Similar Modules
References
N/A
Information for Developers
| Section under construction. |