Documentation/Nightly/Modules/BrainLogisticSegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: Brain Tissues Extension | |||||||
|
Module Description
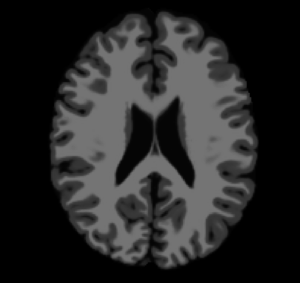
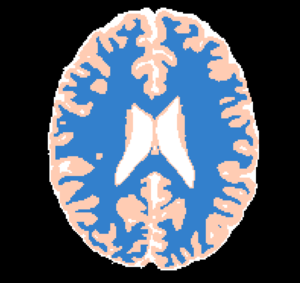
This CLI module offers the Brain Logistic Segmentation (BLS) algorithm, which is a fully automatic segmentation method for MRI structural images. Although the method was intensively tested over T1 weighted (T1w) MRI images, it could be also applied to similar structural MRI data such as T2w and PDw images. The only requirement that is needed in BLS method is that the image histogram can be enough information to describe each tissue of interest (defined as distinguished peaks at image histogram). Full description of BLS method can be found at paper publish at ... journal (DOI: ).
NOTE: A small set of pre-processing steps are usually required for a smooth tissue segmentation using BLS. Methods such as image denoising (e.g. Anomalous Filters) and bias field correction (N4 ITK Bias Field Correction) can greatly assist to the BLS convergence.
Use Cases
- Use Case 1: General brain tissue segmentation
- There are several image quantitative approaches that use only a certain tissue type (for instance, cortical thickness) in which a previous brain segmentation could be needed.
- Use Case 2: Brain extraction refinement
- In some occasions, the brain extraction procedure may left non-brain tissues on the final brain mask (e.g. dura). Using a simple brain tissue mask offered in BLS method can be helpful to remove undesired non-brian information on further analysis (e.g. cortical thickness, total intracranial volume and brain atrophy).
Panels and their use
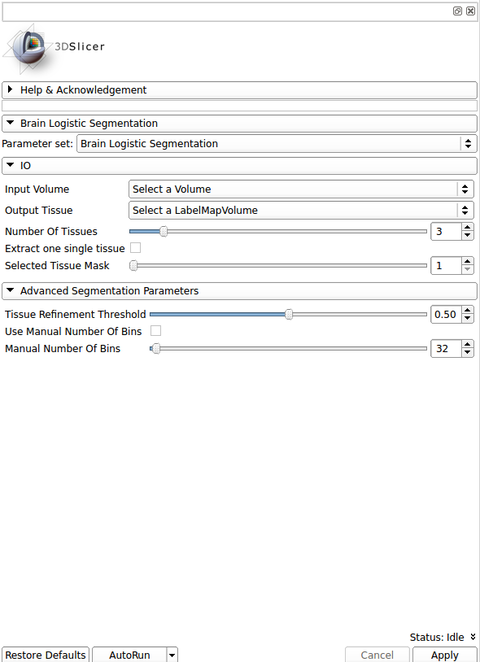
IO:
- Input Volume
- Pick the input to the algorithm. This should be an MRI strutural images with a type listed in the Image Modality option
- Image Modality
- MRI strutural image inserted as a input volume
- Output Volume
- Pick the output to the algorithm (a label image)
Tissue Segmentation Parameters:
- Separate one tissue?
- Select one tissue type desired to be passed as the output. If checked, the tissue type in Tissue Type option is used
- Tissue Type
- Tissue type that will be resulted from the brain segmentation
Similar Modules
References
- paper
Information for Developers
| Section under construction. |