Difference between revisions of "Documentation/Nightly/Modules/BrainExtractionTool"
Acsenrafilho (talk | contribs) m |
Acsenrafilho (talk | contribs) m |
||
| Line 50: | Line 50: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
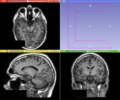
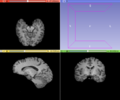
| − | [[Image: | + | [[Image:BrainExtractionTool-GUI.png|thumb|380px|User Interface]] |
*Input Volume: | *Input Volume: | ||
**Insert an input scalar volume | **Insert an input scalar volume | ||
Revision as of 01:02, 26 November 2016
Home < Documentation < Nightly < Modules < BrainExtractionTool
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
This work was partially funded by CAPES and CNPq (grant 201871/2015-7/SWE), a Brazilian research support agency. Information on CAPES can be obtained from the official websites, CAPES here and CAPES here. | |||||||||
|
Module Description
This module aims to extracted the brain in structural MRI images, namely T1 weighted MRI images. The method is based on the non-parametric algorithm proposed by Iglesias et. al. [1]. The application with T2 and proton density MRI images were not intensively tested, but it is assumed that the algorithm is robust with those image contrasts as well. For more details about the brain extraction method, please check the original paper
In addition, for better brain volume extraction results it could be applied some preprocessing steps such as a noise attenuation and field inhomogeneity correction methods in advance. See the modules AAD Image Filter and N4 Bias Field Correction.
Use Cases
Most frequently used for these scenarios:
- Use Case 1: Initial image processing step for many neuroimage analysis, such as in DTI and cortical thickness estimation.
- It would be useful for tissue classication.
- Use Case 2: Information reduction to registration pipelines
- The non-brain tissues tend to decrease the quality of many registration algorithms.
- Use Case 3: Increase the precision of classification algorithms for neurodegenerative brain diseases diagnosis.
- Could offer a better segmentation and classification on specific brain image analysis such as in Multiple Sclerosis lesion segmentation as well as the global brain atrophy.
Tutorials
N/A
Panels and their use
- Input Volume:
- Insert an input scalar volume
- Output Volume:
- Set the output volume
Similar Modules
References
Repositories:
- Source code: GitHub repository
- Issue tracker: open issues and enhancement requests
- ↑ Iglesias, J.E. et al., 2011. "Robust Brain Extraction Across Datasets and Comparison With Publicly Available Methods". IEEE Transactions on Medical Imaging, 30(9), pp.1617–1634