Difference between revisions of "Documentation/Nightly/Modules/BodyCentricCubicMesh"
(changed logo) |
|||
| (22 intermediate revisions by 3 users not shown) | |||
| Line 8: | Line 8: | ||
{{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | {{documentation/{{documentation/version}}/module-introduction-start|{{documentation/modulename}}}} | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
| − | Extension: [[Documentation/{{documentation/version}}/Extensions/ | + | Extension: [[Documentation/{{documentation/version}}/Extensions/CBC_3D_I2MConversion |CBC 3D I2MConversion]]<br> |
Acknowledgments: | Acknowledgments: | ||
| − | *This work was partially supported by NIH R44 OD018334-03A, grant CCF- | + | *This work was partially supported by NIH R44 OD018334-03A, grant CCF-1439079 and by the Richard T.Cheng Endowment. |
| + | |||
| + | Author: Fotis Drakopoulos<br> | ||
| + | Contributors: Joi Best (CRTC), Fotis Drakopoulos (CRTC), Yixun Liu (CRTC), Andrey Fedorov (CRTC), Nikos Chrisochoides (CRTC)<br> | ||
| + | Contact: Nikos Chrisochoides: <email>npchris@gmail.com</email> , Joi Best: <email>jbest002@odu.edu</email><br> | ||
| + | Website: https://crtc.cs.odu.edu | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
License: BSD<br> | License: BSD<br> | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |Image: | + | |Image:Crtc logo.png|Center for Real-time Computing |
}} | }} | ||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
| Line 36: | Line 36: | ||
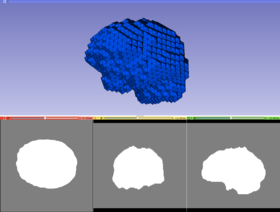
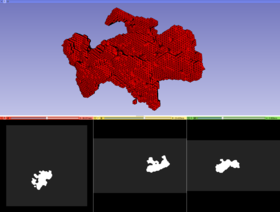
|[[File:ventricles.jpg|thumb|280px|Ventricles BCC mesh]] | |[[File:ventricles.jpg|thumb|280px|Ventricles BCC mesh]] | ||
|} | |} | ||
| − | + | {| | |
| + | |[[File:brain_section.jpg|thumb|280px|Brain BCC mesh cross-section]] | ||
| + | |[[File:nidus_section.jpg|thumb|280px|Nidus BCC mesh cross-section]] | ||
| + | |[[File:ventricles_section.jpg|thumb|280px|Ventricles BCC mesh cross-section]] | ||
| + | |} | ||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| + | N/A | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 43: | Line 50: | ||
{| | {| | ||
| | | | ||
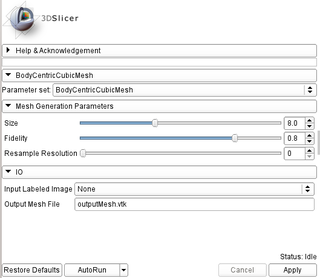
| − | * ''' | + | * '''Mesh Generation Parameters:''' |
| − | ** ''' | + | ** '''Size:''' Controls the size of the mesh. The smaller the value the larger the mesh. Value should be between [1,20] (default: 8). |
| − | |[[Image:BCCPanel.jpg|thumb| | + | ** '''Fidelity:''' Controls the subdivision of the elements that belong to more than one image labels. The larger the value the better the conformity of the mesh on the image label boundaries. Value should be between [0.1,1] (default: 0.8). |
| + | ** '''Resample Resolution:''' Controls the image resampling. Suggested value is 0 (no-resampling) for large tissues and 1 or 2 for smaller tissues. Value should be 0 or 1 or 2 (default: 0). | ||
| + | * '''Input/Output:''' | ||
| + | ** '''Input Labeled Image:''' The input labeled image. | ||
| + | ** '''Output Mesh File:''' The output tetrahedral mesh in vtk file format. (default: outputMesh.vtk) | ||
| + | |[[Image:BCCPanel.jpg|thumb|320px|User Interface]] | ||
|} | |} | ||
| + | '''Note: this module will only work if the input image has identity orientation!''' | ||
| + | |||
| + | <!-- ---------------------------- --> | ||
| + | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| + | *[[Documentation/Nightly/Modules/MeshCompression]] | ||
Latest revision as of 14:08, 7 August 2018
Home < Documentation < Nightly < Modules < BodyCentricCubicMesh
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: CBC 3D I2MConversion
Author: Fotis Drakopoulos | |||
|
Module Description
The module generates a Body Centric Cubic (BCC) mesh from a labeled image. Initially the generated mesh is homogeneous, that means does not distinguish different tissues. Later the module specifies which tissue each tetrahedron belongs to. Each tissue is capable of automatically adjusting its resolution based on its geometric complexity and the predefined subdivision criterion. The current version of the module supports a single-tissue input labeled image.
Use Cases
Tutorials
N/A
Panels and their use
|
Note: this module will only work if the input image has identity orientation!
Similar Modules
References
- Fedorov A., Chrisochoides N., Kikinis R., Warfield S, "Tetrahedral mesh generation for medical imaging", The Insight Journal-2005 MICCAI Open-Source Workshop, 2005
- Yixun Liu, Panagiotis Foteinos, Andrey Chernikov and Nikos Chrisochoides, "Mesh Deformation-based Multi-tissue Mesh Generation for Brain Images", Engineering with Computers, Volume 28, pages 305-318, 2012.
Information for Developers
| Section under construction. |