Difference between revisions of "Documentation/Nightly/Modules/BasicBrainTissues"
Acsenrafilho (talk | contribs) (BasicBrainTissue wiki page created) |
Acsenrafilho (talk | contribs) |
||
| Line 22: | Line 22: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | This module offer a... | + | This module offer a simple and robust brain tissue segmentation focused on white matter, gray matter and CSF brain tissues. The method applied here is based on K-Means clustering segmentation, which presents best results with high quality T1 weighted MRI images. |
| + | |||
| + | '''NOTE''': This module can be used alone (only apply the K-Means segmentation method on the input data), but the [[Documentation/{{documentation/version}}/Modules/BrainStructuresSegmenter|Brain Structures Segmenter]] module already use it internally, which control a image processing pipeline for a better brain tissue segmentation result. | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | * Use Case 1: a | + | * Use Case 1: Separate White Matter, Gray Matter and CSF brain tissues from a strucutral MRI image. |
| − | ** | + | **There are some image processing strategies that may need a specific brain tissue mask and this module could facilitate this task. For instance, a Multiple Sclerosis lesion detection algorithm may need a White Matter mask in order to define a localized brain region where the lesion are more probable to appear. |
<gallery widths="200px" perrow="3"> | <gallery widths="200px" perrow="3"> | ||
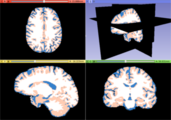
| − | Image: | + | Image:T1_tissues.png|White matter, gray matter and CSF tissues segmented from the previous MRI image |
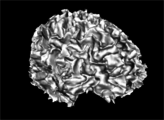
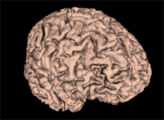
| − | Image: | + | Image:WM_3DReconstruction.png|A white matter mask 3D reconstruction |
| + | Image:GM_3DReconstruction.png|A gray matter mask 3D reconstruction | ||
</gallery> | </gallery> | ||
| Line 37: | Line 40: | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
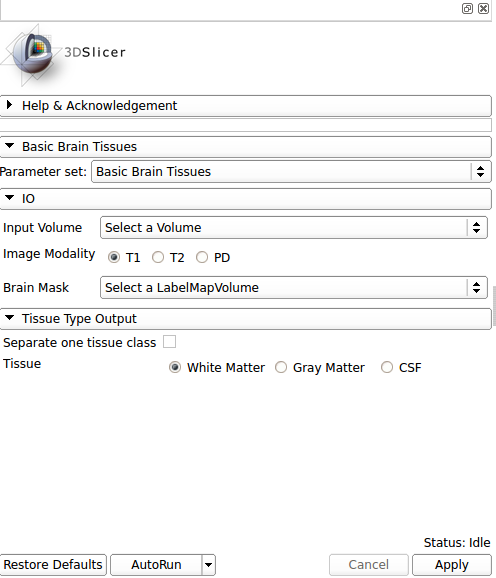
| − | [[Image: | + | [[Image:basicbraintissues_gui.png|thumb|500px|User Interface]] |
IO: | IO: | ||
*Input Volume | *Input Volume | ||
| − | **Select the input | + | **Input volume. The algorithm works better with high resolution T1 MRI images alread brain extract and inhomogeneity corrected |
| − | * | + | *Image Modality |
| − | ** | + | **Select the image modality inserted as a input volume |
| + | *Brain Mask | ||
| + | **Output brain tissue mask | ||
| − | + | Tissue Type Output: | |
| − | * | + | *Separate one tissue class |
| − | ** | + | **Choose if you want all the tissues classes or only one class segmented |
| − | * | + | *Tissue |
| − | ** | + | **Choose what is the brain tissue label that you want as the output label |
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | *[ | + | *[https://www.slicer.org/wiki/Modules:EMSegmenter-3.6 EM Segmenter] |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | + | N/A | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 14:02, 27 November 2016
Home < Documentation < Nightly < Modules < BasicBrainTissues
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: BrainTissuesExtension | |||||||
|
Module Description
This module offer a simple and robust brain tissue segmentation focused on white matter, gray matter and CSF brain tissues. The method applied here is based on K-Means clustering segmentation, which presents best results with high quality T1 weighted MRI images.
NOTE: This module can be used alone (only apply the K-Means segmentation method on the input data), but the Brain Structures Segmenter module already use it internally, which control a image processing pipeline for a better brain tissue segmentation result.
Use Cases
- Use Case 1: Separate White Matter, Gray Matter and CSF brain tissues from a strucutral MRI image.
- There are some image processing strategies that may need a specific brain tissue mask and this module could facilitate this task. For instance, a Multiple Sclerosis lesion detection algorithm may need a White Matter mask in order to define a localized brain region where the lesion are more probable to appear.
Panels and their use
IO:
- Input Volume
- Input volume. The algorithm works better with high resolution T1 MRI images alread brain extract and inhomogeneity corrected
- Image Modality
- Select the image modality inserted as a input volume
- Brain Mask
- Output brain tissue mask
Tissue Type Output:
- Separate one tissue class
- Choose if you want all the tissues classes or only one class segmented
- Tissue
- Choose what is the brain tissue label that you want as the output label
Similar Modules
References
N/A
Information for Developers
| Section under construction. |