Difference between revisions of "Documentation/Nightly/Modules/AirwaySegmentation"
(Created page with '<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- -----------------------…') |
m (Updating contributor's emails) |
||
| (39 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
Extension: [[Documentation/{{documentation/version}}/Extensions/AirwaySegmentation|AirwaySegmentation]]<br> | Extension: [[Documentation/{{documentation/version}}/Extensions/AirwaySegmentation|AirwaySegmentation]]<br> | ||
| − | This work is | + | Acknowledgments: |
| − | Author: Pietro Nardelli | + | This work is supported by NA-MIC, the Slicer Community and University College of Cork.<br> |
| − | Contributor1: | + | Author: Pietro Nardelli (University College Cork)<br> |
| − | Contact: Pietro Nardelli, <email> | + | Contributor1:<br> |
| + | Contact: Pietro Nardelli, <email>pnardelli@bwh.harvard.edu</email>, <email>pie.nardelli@gmail.com</email><br> | ||
{{documentation/{{documentation/version}}/module-introduction-row}} | {{documentation/{{documentation/version}}/module-introduction-row}} | ||
{{documentation/{{documentation/version}}/module-introduction-logo-gallery | {{documentation/{{documentation/version}}/module-introduction-logo-gallery | ||
| − | |{{collaborator|logo| | + | |{{collaborator|logo|namic}}|{{collaborator|longname|namic}} |
| − | |{{collaborator|logo|spl}}|{{collaborator|longname|spl}} | + | |Image:UCC-logoNEW.png|University College Cork (UCC) |
| + | |Image:Biolectromagnetics_group_UCC.jpg|Bioelectromagnetics group UCC | ||
| + | |{{collaborator|logo|spl}}|{{collaborator|longname|spl}} | ||
}} | }} | ||
| + | |||
{{documentation/{{documentation/version}}/module-introduction-end}} | {{documentation/{{documentation/version}}/module-introduction-end}} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Module Description}} | {{documentation/{{documentation/version}}/module-section|Module Description}} | ||
| − | + | AirwaySegmentation is an extension for airway segmentation starting from chest CT images. Two modules come with this extension. A simple CLI module, called Airway Segmentation CLI, that uses a modified version of ITK's itkConnectedThresholdImageFilter to segment all the pixels with an intensity below a threshold. The threshold is automatically identified by the module. To use this module for the segmentation, the user is required to place only one | |
| − | + | fiducial point within the trachea. This fiducial is used as starting point for the region growing segmentation. Also, using this module, the user needs to specify the convolution kernel used for reconstructing the DICOM image. A scripted module, called AirwaySegmentation, is also available. This is a python module that automatically invokes the CLI module described above. It automatically extracts the convolution kernel of the image, and for this reason it only works for DICOM images. The output volume is also automatically created. This way, the user needs to only specify the input volume, and the fiducial placed in the trachea. A label color can also be optionally selected. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
| − | + | *Airway Segmentation starting from chest CT datasets | |
| + | |||
| + | {| | ||
| + | |[[Image:Screenshot from 2015-01-08 11-32-48.jpg|thumb|500px|Airway Segmentation Scripted Module: example of usage]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Tutorials}} | {{documentation/{{documentation/version}}/module-section|Tutorials}} | ||
| + | |||
N/A | N/A | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Panels and their use}} | {{documentation/{{documentation/version}}/module-section|Panels and their use}} | ||
| − | + | ||
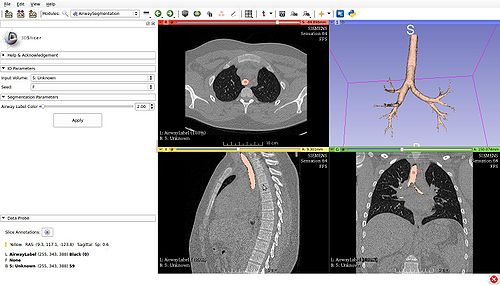
| − | < | + | * <span>'''''IO Parameters'''''</span>: Input and Output parameters. |
| − | { | + | ** <span>'''Input Volume'''</span>: Input chest CT dataset to be segmented. |
| − | + | ** <span>'''Seed'''</span>: Seed point for the algorithm. Only one seed point must be placed within the trachea. If using an pig CT chest image, the fiducial has to be placed between the carina and the further branch coming out of the trachea. | |
| + | |||
| + | * <span>'''''Segmentation Parameters'''''</span>: Input parameters for segmentation (optional). | ||
| + | ** <span>'''Airway Label Color'''</span>: The integer value (0-255) to use for the segmentation results. This will determine the color of the segmentation that will be generated by the algorithm. | ||
| + | The user interface panel for the scripted module: | ||
| + | |||
| + | {| | ||
| + | |[[Image:AirwaySegmentation_Scripted_Interface.jpg|thumb|300px|{{documentation/modulename}} Scripted Module Interface ]] | ||
| + | |} | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
| Line 52: | Line 63: | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | N | + | Nardelli, P., Khan, K. A., Corvò, A., Moore, N., Murphy, M. J., Twomey, M., O’Connor, O. J., Kennedy, M. P., Estépar, R. S. J., Maher, M. M. & Cantillon-Murphy, P. (2015). Optimizing parameters of an open-source airway segmentation algorithm using different CT images. Biomedical engineering online, 14(1), 62. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Information for Developers}} | {{documentation/{{documentation/version}}/module-section|Information for Developers}} | ||
| − | + | The code is available at [https://github.com/PietroNardelli/Slicer-AirwaySegmentation Github]. | |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-footer}} | {{documentation/{{documentation/version}}/module-footer}} | ||
| + | [[Category:Documentation/{{documentation/version}}/Modules/Segmentation]] | ||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 09:29, 3 January 2018
Home < Documentation < Nightly < Modules < AirwaySegmentation
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: AirwaySegmentation | |||||||||
|
Module Description
AirwaySegmentation is an extension for airway segmentation starting from chest CT images. Two modules come with this extension. A simple CLI module, called Airway Segmentation CLI, that uses a modified version of ITK's itkConnectedThresholdImageFilter to segment all the pixels with an intensity below a threshold. The threshold is automatically identified by the module. To use this module for the segmentation, the user is required to place only one fiducial point within the trachea. This fiducial is used as starting point for the region growing segmentation. Also, using this module, the user needs to specify the convolution kernel used for reconstructing the DICOM image. A scripted module, called AirwaySegmentation, is also available. This is a python module that automatically invokes the CLI module described above. It automatically extracts the convolution kernel of the image, and for this reason it only works for DICOM images. The output volume is also automatically created. This way, the user needs to only specify the input volume, and the fiducial placed in the trachea. A label color can also be optionally selected.
Use Cases
- Airway Segmentation starting from chest CT datasets
Tutorials
N/A
Panels and their use
- IO Parameters: Input and Output parameters.
- Input Volume: Input chest CT dataset to be segmented.
- Seed: Seed point for the algorithm. Only one seed point must be placed within the trachea. If using an pig CT chest image, the fiducial has to be placed between the carina and the further branch coming out of the trachea.
- Segmentation Parameters: Input parameters for segmentation (optional).
- Airway Label Color: The integer value (0-255) to use for the segmentation results. This will determine the color of the segmentation that will be generated by the algorithm.
The user interface panel for the scripted module:
Similar Modules
N/A
References
Nardelli, P., Khan, K. A., Corvò, A., Moore, N., Murphy, M. J., Twomey, M., O’Connor, O. J., Kennedy, M. P., Estépar, R. S. J., Maher, M. M. & Cantillon-Murphy, P. (2015). Optimizing parameters of an open-source airway segmentation algorithm using different CT images. Biomedical engineering online, 14(1), 62.
Information for Developers
The code is available at Github.