Difference between revisions of "Documentation/Nightly/Modules/AFTSegmenter"
From Slicer Wiki
Acsenrafilho (talk | contribs) (Created page with "<noinclude>{{documentation/versioncheck}}</noinclude> <!-- ---------------------------- --> {{documentation/{{documentation/version}}/module-header}} <!-- --------------------...") |
Acsenrafilho (talk | contribs) |
||
| Line 24: | Line 24: | ||
[[File:AFTSegmenter-icon.png|right]] | [[File:AFTSegmenter-icon.png|right]] | ||
| − | This module offers ... | + | This module offers an implementation of a recent Multiple Sclerosis lesion segmentation approach based on a unsupervised method described by Cabezas et al. <ref>Cabezas M. et al.(2014) "Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding", Computer Methods and Programs in Biomedicine, DOI: 10.1016/j.cmpb.2014.04.006</ref>. This module is intended to be used with FLAIR and T1 MRI volumes, which the MS lesions can be detected. |
| Line 30: | Line 30: | ||
{{documentation/{{documentation/version}}/module-section|Use Cases}} | {{documentation/{{documentation/version}}/module-section|Use Cases}} | ||
* Use Case 1: Multiple Sclerosis (MS) lesions segmentation | * Use Case 1: Multiple Sclerosis (MS) lesions segmentation | ||
| − | **... | + | **Using T1 and FLAIR MRI volumes, it can be possible to detect abnormal voxel signal using a parametric strategy, which delineates white matter signals that does not belongs to the majority neighborhood pattern. More details can be found in the original paper <ref>Cabezas M. et al.(2014) "Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding", Computer Methods and Programs in Biomedicine, DOI: 10.1016/j.cmpb.2014.04.006</ref> |
<gallery widths="300px" heights="300px" perrow="3"> | <gallery widths="300px" heights="300px" perrow="3"> | ||
| Line 44: | Line 44: | ||
IO: | IO: | ||
*T1 Volume | *T1 Volume | ||
| + | **Input T1 volume | ||
*T2-FLAIR Volume | *T2-FLAIR Volume | ||
| + | **Input T2-FLAIR volume | ||
*Lesion Label | *Lesion Label | ||
**Output a global lesion mask | **Output a global lesion mask | ||
| − | + | *Is brain extracted? | |
| − | + | **Is the input data (T1 and T2-FLAIR) already brain extracted? | |
| − | * | ||
| − | ** | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Segmentation Parameters: | Segmentation Parameters: | ||
| Line 70: | Line 58: | ||
**Define the outlier detection based on units of standard deviation in the T2-FLAIR gray matter voxel intensity distribution | **Define the outlier detection based on units of standard deviation in the T2-FLAIR gray matter voxel intensity distribution | ||
*White Matter Matching | *White Matter Matching | ||
| − | **Set the local neighborhood searching for label refinement step. This metric defines the percentage of white matter tissue that surrounds the hyperintense lesions. | + | **Set the local neighborhood searching for label refinement step. This metric defines the percentage of white matter tissue that surrounds the hyperintense lesions. Large values defines a conservative segmentation, i.e. in order to define a true MS lesion, it must be close to certain percentage of white matter area. |
*Minimum Lesion Size | *Minimum Lesion Size | ||
**Set the minimum lesion size adopted as a true lesion in the final lesion map. Units are given in number of voxels | **Set the minimum lesion size adopted as a true lesion in the final lesion map. Units are given in number of voxels | ||
*Gray Matter Mask Value | *Gray Matter Mask Value | ||
| − | **Set the mask value that represents the gray matter. Default is defined based on the Basic Brain Tissues | + | **Set the mask value that represents the gray matter. Default is defined based on the ([https://www.slicer.org/wiki/Documentation/Nightly/Extensions/BrainTissuesExtension Basic Brain Tissues] extension) output |
*White Matter Mask Value | *White Matter Mask Value | ||
| − | **Set the mask value that represents the white matter. Default is defined based on the Basic Brain Tissues | + | **Set the mask value that represents the white matter. Default is defined based on the ([https://www.slicer.org/wiki/Documentation/Nightly/Extensions/BrainTissuesExtension Basic Brain Tissues] extension) output |
| − | |||
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|Similar Modules}} | {{documentation/{{documentation/version}}/module-section|Similar Modules}} | ||
| − | + | *[[Documentation/Nightly/Modules/LSSegmenter|LS Segmenter]] | |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
{{documentation/{{documentation/version}}/module-section|References}} | {{documentation/{{documentation/version}}/module-section|References}} | ||
| − | * | + | * Cabezas, M., Oliver, A., Roura, E., Freixenet, J., Vilanova, J. C., Ramió-Torrentà, L., Rovira, À. and Lladó, X. (2014) ‘Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding’, Computer Methods and Programs in Biomedicine. Elsevier Ireland Ltd, 115(3), pp. 147–161. DOI: 10.1016/j.cmpb.2014.04.006. |
<!-- ---------------------------- --> | <!-- ---------------------------- --> | ||
Revision as of 19:52, 24 July 2017
Home < Documentation < Nightly < Modules < AFTSegmenter
|
For the latest Slicer documentation, visit the read-the-docs. |
Introduction and Acknowledgements
|
Extension: LesionSpotlight | |||||||
|
Module Description
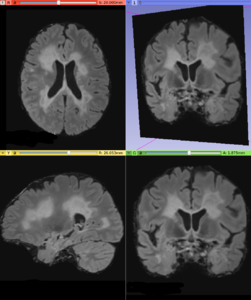
This module offers an implementation of a recent Multiple Sclerosis lesion segmentation approach based on a unsupervised method described by Cabezas et al. [1]. This module is intended to be used with FLAIR and T1 MRI volumes, which the MS lesions can be detected.
Use Cases
- Use Case 1: Multiple Sclerosis (MS) lesions segmentation
- Using T1 and FLAIR MRI volumes, it can be possible to detect abnormal voxel signal using a parametric strategy, which delineates white matter signals that does not belongs to the majority neighborhood pattern. More details can be found in the original paper [2]
Panels and their use
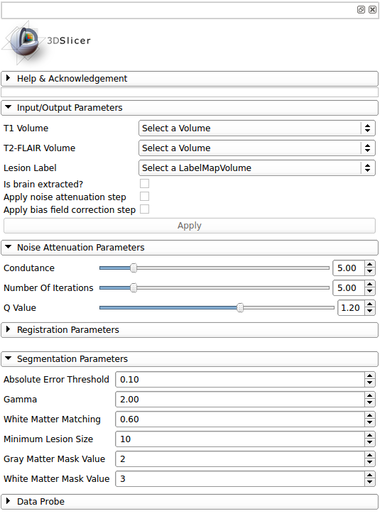
IO:
- T1 Volume
- Input T1 volume
- T2-FLAIR Volume
- Input T2-FLAIR volume
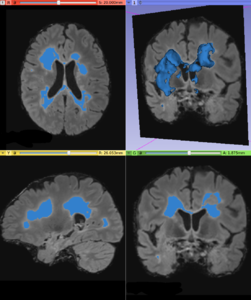
- Lesion Label
- Output a global lesion mask
- Is brain extracted?
- Is the input data (T1 and T2-FLAIR) already brain extracted?
Segmentation Parameters:
- Absolute Error Threshold
- Define the absolute error threshold for gray matter statistics. This measure evaluated the similarity between the MNI152 template and the T2-FLAIR gray matter fluctuation estimative. A higher error gives a higher variability in the final lesion segmentation
- Gamma
- Define the outlier detection based on units of standard deviation in the T2-FLAIR gray matter voxel intensity distribution
- White Matter Matching
- Set the local neighborhood searching for label refinement step. This metric defines the percentage of white matter tissue that surrounds the hyperintense lesions. Large values defines a conservative segmentation, i.e. in order to define a true MS lesion, it must be close to certain percentage of white matter area.
- Minimum Lesion Size
- Set the minimum lesion size adopted as a true lesion in the final lesion map. Units are given in number of voxels
- Gray Matter Mask Value
- Set the mask value that represents the gray matter. Default is defined based on the (Basic Brain Tissues extension) output
- White Matter Mask Value
- Set the mask value that represents the white matter. Default is defined based on the (Basic Brain Tissues extension) output
Similar Modules
References
- Cabezas, M., Oliver, A., Roura, E., Freixenet, J., Vilanova, J. C., Ramió-Torrentà, L., Rovira, À. and Lladó, X. (2014) ‘Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding’, Computer Methods and Programs in Biomedicine. Elsevier Ireland Ltd, 115(3), pp. 147–161. DOI: 10.1016/j.cmpb.2014.04.006.
Information for Developers
| Section under construction. |
- ↑ Cabezas M. et al.(2014) "Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding", Computer Methods and Programs in Biomedicine, DOI: 10.1016/j.cmpb.2014.04.006
- ↑ Cabezas M. et al.(2014) "Automatic multiple sclerosis lesion detection in brain MRI by FLAIR thresholding", Computer Methods and Programs in Biomedicine, DOI: 10.1016/j.cmpb.2014.04.006